













| General Information: |
|
| Name(s) found: |
sc-PA /
FBpp0070072
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 345 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
|
| Biological Process: |
nervous system development
[IGI][TAS]
neuroblast fate determination [NAS][TAS] peripheral nervous system development [IMP][NAS][TAS] regulation of transcription, DNA-dependent [NAS] sex determination, establishment of X:A ratio [NAS][TAS] sex determination [IGI][IMP][NAS][TAS] regulation of transcription [IEA] regulation of mitotic cell cycle [TAS] sensory organ development [IMP] regulation of transcription from RNA polymerase II promoter [ISS] ventral cord development [NAS] bristle morphogenesis [IMP][NAS] central nervous system development [NAS][TAS] dosage compensation [NAS] negative regulation of apoptosis [IMP] |
| Molecular Function: |
DNA binding
[NAS][TAS]
protein heterodimerization activity [IPI] specific RNA polymerase II transcription factor activity [ISS] sequence-specific DNA binding [IDA] transcription activator activity [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKNNNNTTKS TTMSSSVLST NETFPTTINS ATKIFRYQHI MPAPSPLIPG GNQNQPAGTM 60
61 PIKTRKYTPR GMALTRCSES VSSLSPGSSP APYNVDQSQS VQRRNARERN RVKQVNNSFA 120
121 RLRQHIPQSI ITDLTKGGGR GPHKKISKVD TLRIAVEYIR RLQDLVDDLN GGSNIGANNA 180
181 VTQLQLCLDE SSSHSSSSST CSSSGHNTYY QNTISVSPLQ QQQQLQRQQF NHQPLTALSL 240
241 NTNLVGTSVP GGDAGCVSTS KNQQTCHSPT SSFNSSMSFD SGTYEGVPQQ ISTHLDRLDH 300
301 LDNELHTHSQ LQLKFEPYEH FQLDEEDCTP DDEEILDYIS LWQEQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
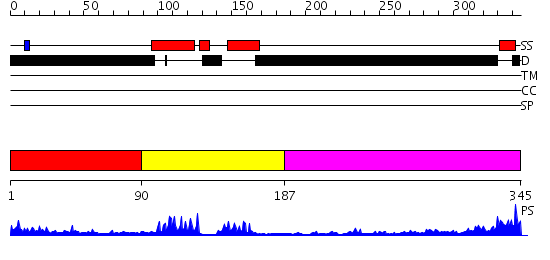
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [90..186] | 12.30103 | SREBP-1a | |
| 3 | View Details | [187..345] | 2.296991 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)