













| General Information: |
|
| Name(s) found: |
CaMKI-PB /
FBpp0088138
[FlyBase]
CaMKI-PA / FBpp0088136 [FlyBase] CaMKI-PG / FBpp0088134 [FlyBase] |
| Description(s) found:
Found 66 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 405 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[ISS]
|
| Biological Process: |
protein amino acid phosphorylation
[IEA][ISS][NAS]
|
| Molecular Function: |
ATP binding
[IEA]
calmodulin binding [TAS] calmodulin-dependent protein kinase activity [ISS] protein serine/threonine kinase activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLFGKKDSG KKAKAKDLKE LNKQVSIEEK YNLHGLLGTG AFSEVRLAES KDSPGEHFAV 60
61 KIIDKKALKG KEESLENEIR VLRRFSANHF DGKCLNGTRL THPNIVQLLE TYEDKSKVYL 120
121 VMELVTGGEL FDRIVEKGSY TEKDASHLIR QILEAVDYMH EQGVVHRDLK PENLLYYSPD 180
181 DDSKIMISDF GLSKMEDSGI MATACGTPGY VAPEVLAQKP YGKAVDVWSI GVISYILLCG 240
241 YPPFYDENDA NLFAQILKGD FEFDSPYWDE ISESAKHFIK NLMCVTVEKR YTCKQALGHA 300
301 WISGNEASSR NIHGTVSEQL KKNFAKSRWK QAYYAATVIR QMQRMALNSN SNANFDSSNS 360
361 SNQDSTTPTA ATGAWTSNVL SSQQSVQSHA QEMNKSGGST NAASQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
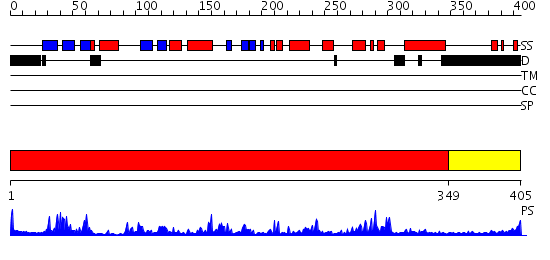
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..348] | 84.39794 | CALMODULIN-DEPENDENT PROTEIN KINASE FROM RAT | |
| 2 | View Details | [349..405] | 9.045757 | Regulatory apparatus of Calcium Dependent protein kinase from Arabidopsis thaliana |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)