













| General Information: |
|
| Name(s) found: |
FBpp0306618
[FlyBase]
Gcn2-PA / FBpp0085148 [FlyBase] |
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1589 amino acids |
Gene Ontology: |
|
| Cellular Component: |
catalytic step 2 spliceosome
[IDA]
precatalytic spliceosome [IDA] cytosol [IC] |
| Biological Process: |
regulation of translation
[IC]
protein amino acid phosphorylation [IDA][NAS] nuclear mRNA splicing, via spliceosome [IC] |
| Molecular Function: |
ATP binding
[IEA]
protein kinase activity [NAS] elongation factor-2 kinase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADEKAKESF RERQAQELEV IKSIFGCDVE DLRPQANPSL WKPTDIRIQL TPLRDSSNGL 60
61 ETYVCTKLHV TCPSKYPKLP PKISLEESKG MSDQLLEALR NQLQAQSQEL RGEVMIYELA 120
121 QTVQAFLLEH NKPPKGSFYD QMLQDKQKRD QELQDIQRQR ESLQRQTLID EVERRKEMFK 180
181 TEAKRRGEPR RSMSESNPRH PSSSESSENS SPYYRGHIYP SKCLDHRNTE TLYFHKMGRQ 240
241 IQRGCCVGHS QRGCIAYTGI DMHCGQLLYI TEWNIKYSQL EQPCIGGGKC HWSSESKCMG 300
301 SHRVDEVMAS IEKQVSSLSQ LQHKNLVSYE CVLCIKRKEG LLVYLVQDFL LGTSVFSISS 360
361 SLGWCMDGAR MVARGVLDAL VFLHNKGVSH SHLLDTTVFM DNTGNVRVSD FSLVPNLLEL 420
421 LSGAGQSSSC GDLPALGALV ESLMPTNSYE MRDFVDKCNS DRTLSASELL EHPFLRFYVD 480
481 NGQQQVMPLP QQQHPNTVQR TGSAMPYQIP TLALSQSRLR TEFEVLMYLG KGAFGDVLKV 540
541 RNILDNREYA IKRIPLPARS RQLYKKMTRE VELLSRLNHE NVVRYFNSWI ESVDDADAAE 600
601 MDKLLGGEWS QSQQDLSVKP AKSPQLGPTL EEDEDEEDSS SSMWNGYIPN MEDSDSDGIE 660
661 FVDSNGKVAV YDDEEQEDST RGKTNSPKPL MQVMYIQMEF CEKCTLRTAI DDNLFNDTDR 720
721 LWRLFREIAE GLAHIHQQGI IHRDLKPVNI FLDSHDQIKI GDFGLATTSF LALQAHDAAP 780
781 APVNQITSAE DGTGTGKVGT TLYVAPELTG NASKSVYNQK VDMYTLGIIL FEMCQPPFDT 840
841 SMERAQTIMA LRNVSINIPD AMLKDPKYEK TVKMLQWLLN HDPAQRPTAE ELLISDLVPP 900
901 AQLEANELQE MLRHALANPQ SKAYKNLVAR CLQQESDEVL EHTYHLGSSR AMKSWNSAII 960
961 IDDIVSLNPV IEFVKAKVVN LFRKHGAIEV DSPLLSPLSA RNSTANANAN AVHLMTHSGC1020
1021 VVVLPCDLRT QFARHVTMNS VNLIRRYCVD RVYREERVFN FHPKQSYDCS FDIIAPTTGS1080
1081 HLVDAELLSL AFEITSELPR LREKNLAIRM NHTNLLRAIL IFCNVPKAQY GALFEGTMDF1140
1141 IESRISRFQF HSSITGIMEK SRTSAQTLMD MLLANFLLTG SRSTVDDSAL KSLMRGKGEA1200
1201 ASLARGALRE LETVVGLAYS LGVKCPIHIW AGLPISFDRA SNGGIVWQMT ADLKPNRSGH1260
1261 PSVLAIGERY DSMLHEFQKQ AQKFNPAMPA RGVLSGAGLT FSLDKLVAAV GVEYAKDCRA1320
1321 IDVGICVCGT RPPLKDVTYI MRLLWSVGIR CGIVEAASEL GDEAQDLARL GALHVILVAE1380
1381 NGSLRVRSFE RERFQERHLT RTELVEFIQK MLRSDGLNGT TVDNFSQLSA LGSGDNRSSG1440
1441 GKERERGENG LSTSASNATI KNNYSQLPNL QVTFLTHDKP TANYKRRLEN QVAQQMSSTL1500
1501 SQFLKKETFV VLVVELPPAV VNAIVGAINP REIRKRETEP EINYVIERFS KYKRYISEIN1560
1561 EEVVDYLSDA KTPIVALYSI SDSYYRVII |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
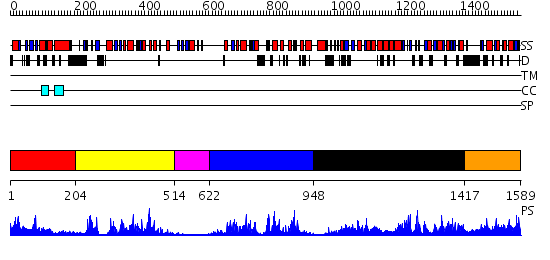
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..203] | 30.39794 | Solution structure of the RWD domain of mouse GCN2 | |
| 2 | View Details | [204..513] | 53.221849 | Aurora-2 T287D T288D complexed with PNU-680632 | |
| 3 | View Details | [514..621] | 5.05 | No description for 2javA was found. | |
| 4 | View Details | [622..947] | 46.154902 | Crystal Structure of eIF2alpha Protein Kinase GCN2: R794G Hyperactivating Mutant in Apo Form. | |
| 5 | View Details | [948..1416] | 97.09691 | Histidyl-tRNA synthetase (HisRS), C-terminal domain; Histidyl-tRNA synthetase (HisRS) | |
| 6 | View Details | [1417..1589] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)