













| General Information: |
|
| Name(s) found: |
tankyrase-PA /
FBpp0084264
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1181 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cytoskeletal anchoring at plasma membrane
[ISS]
|
| Molecular Function: |
structural constituent of cytoskeleton
[ISS]
NAD+ ADP-ribosyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANSSRSRAI LSVNLDAVMA NDPLRELFEA CKTGEIAKVK KLITPQTVNA RDTAGRKSTP 60
61 LHFAAGYGRR EVVEFLLNSG ASIQACDEGG LHPLHNCCSF GHAEVVRLLL KAGASPNTTD 120
121 NWNYTPLHEA ASKGKVDVCL ALLQHGANHT IRNSEQKTPL ELADEATRPV LTGEYRKDEL 180
181 LEAARSGAED RLLALLTPLN VNCHASDGRR STPLHLAAGY NRIGIVEILL ANGADVHAKD 240
241 KGGLVPLHNA CSYGHFDVTK LLIQAGANVN ANDLWAFTPL HEAASKSRVE VCSLLLSRGA 300
301 DPTLLNCHSK SAIDAAPTRE LRERIAFEYK GHCLLDACRK CDVSRAKKLV CAEIVNFVHP 360
361 YTGDTPLHLA VVSPDGKRKQ LMELLTRKGS LLNEKNKAFL TPLHLAAELL HYDAMEVLLK 420
421 QGAKVNALDS LGQTPLHRCA RDEQAVRLLL SYAADTNIVS LEGLTAAQLA SDSVLKLLKN 480
481 PPDSETHLLE AAKAGDLDTV RRIVLNNPIS VNCRDLDGRH STPLHFAAGF NRVPVVQFLL 540
541 EHGAEVYAAD KGGLVPLHNA CSYGHYEVTE LLVKHGANVN VSDLWKFTPL HEAAAKGKYD 600
601 ICKLLLKHGA DPMKKNRDGA TPADLVKESD HDVAELLRGP SALLDAAKKG NLARVQRLVT 660
661 PESINCRDAQ GRNSTPLHLA AGYNNFECAE YLLENGADVN AQDKGGLIPL HNASSYGHLD 720
721 IAALLIKHKT VVNATDKWGF TPLHEAAQKG RTQLCSLLLA HGADAYMKNQ EGQTPIELAT 780
781 ADDVKCLLQD AMATSLSQQA LSASTQSLTS SSPAPDATAA AAPGTSSSSS SAILSPTTET 840
841 VLLPTGASMI LSVPVPLPLS SSTRISPAQG AEANGAEGSS SDDLLPDADT ITNVSGFLSS 900
901 QQLHHLIELF EREQITLDIL AEMGHDDLKQ VGVSAYGFRH KILKGIAQLR STTGIGNNVN 960
961 LCTLLVDLLP DDKEFVAVEE EMQATIREHR DNGQAGGYFT RYNIIRVQKV QNRKLWERYA1020
1021 HRRQEIAEEN FLQSNERMLF HGSPFINAIV QRGFDERHAY IGGMFGAGIY FAEHSSKSNQ1080
1081 YVYGIGGGIG CPSHKDKSCY VCPRQLLLCR VALGKSFLQY SAMKMAHAPP GHHSVVGRPS1140
1141 AGGLHFAEYV VYRGEQSYPE YLITYQIVKP DDSSSGTEDT R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
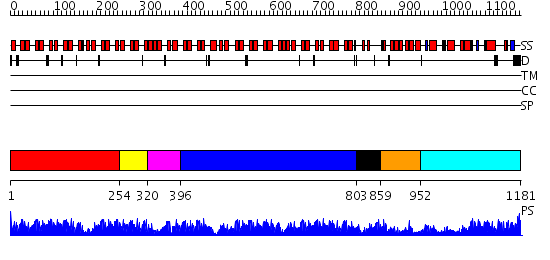
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..253] | 45.69897 | X-ray structure of human gankyrin | |
| 2 | View Details | [254..319] | 45.0 | Crystal structure of a designed selected Ankyrin Repeat protein in complex with the Maltose Binding Protein | |
| 3 | View Details | [320..395] | 13.30103 | Pyk2-associated protein beta; Pyk2-associated protein beta ARF-GAP domain | |
| 4 | View Details | [396..802] | 97.09691 | Ankyrin-R | |
| 5 | View Details | [803..858] | 44.0 | No description for 2dvwA was found. | |
| 6 | View Details | [859..951] | 3.6 | No description for 2e8oA was found. | |
| 7 | View Details | [952..1181] | 35.154902 | No description for 2rf5A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.53 |
Source: Reynolds et al. (2008)