













| General Information: |
|
| Name(s) found: |
Fur1-PA /
FBpp0089025
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1101 amino acids |
Gene Ontology: |
|
| Cellular Component: |
integral to Golgi membrane
[NAS]
integral to membrane [NAS] Golgi stack [NAS] plasma membrane [IDA][ISS] |
| Biological Process: |
proteolysis
[IEA][NAS]
|
| Molecular Function: |
serine-type endopeptidase activity
[IDA][NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKNDVVRWSR QPTSNTTNSS SSSRSDSNST HKHRSKSNKL NARQLGSNAA RSCQQRSSVA 60
61 TTLEDEQQTI IECDIGNFNF DCNLFKTSFL TQHKQKRSGK SSSKSKSNRS RPLAKTKAVF 120
121 LLALQFSAVV FLCNINVGFV AGSVATAASS AGGSSPAAPS SAPSSPPTVA VPPPPPPSSA 180
181 LKVDPNGQSP VLPPYVLDYE TGGKAKLTPN NGKFGQSGSS GSNNNHIVGH YTHTWAVHIP 240
241 NGDNGMADAV AKDHGFVNLG KIFDDHYHFA HHKVSKRSLS PATHHQTRLD DDDRVHWAKQ 300
301 QRAKSRSKRD FIRMRPSRTS SRAMSMVDAM SFNDSKWPQM WYLNRGGGLD MNVIPAWKMG 360
361 ITGKGVVVTI LDDGLESDHP DIQDNYDPKA SYDVNSHDDD PMPHYDMTDS NRHGTRCAGE 420
421 VAATANNSFC AVGIAYGASV GGVRMLDGDV TDAVEARSLS LNPQHIDIYS ASWGPDDDGK 480
481 TVDGPGELAS RAFIEGTTKG RGGKGSIFIW ASGNGGREQD NCNCDGYTNS IWTLSISSAT 540
541 EEGHVPWYSE KCSSTLATTY SSGGQGEKQV VTTDLHHSCT VSHTGTSASA PLAAGIAALV 600
601 LQSNQNLTWR DLQHIVVRTA KPANLKDPSW SRNGVGRRVS HSFGYGLMDA AEMVRVARNW 660
661 KAVPEQQRCE INAPHVDKVI PPRTHITLQL TVNHCRSVNY LEHVQAKITL TSQRRGDIQL 720
721 FLRSPANTSV TLLTPRIHDN SRSGFNQWPF MSVHTWGESP QGNWQLEIHN EGRYMAQITQ 780
781 WDMIFYGTET PAQPDDVANP SQSNQFNLYG NDMAHNDVEY DSTGQWRNMQ QVGEVGMTRD 840
841 HSNTAACLKW SDRKCLECND SAYMFEDQCY DVCPVHTYPL DKFQAEEDEQ DDEVTRGPVN 900
901 PYSSSPMDHS LLMSNSLDDK QDPLQAEDRR RRSSLTQLVE VPSRVCAACD RSCLECYGAL 960
961 ASQCSTCSPG SQLRKILNET FCYAYVVRST GMASVVDISK MDDRDTQQYM TGTTVLLLVS1020
1021 VIFTLMGVAV AGGIVYHRRA MARSNELYSR VSLVPGDESD SDEDELFTAH FPARKSGVNI1080
1081 YRDEAPSEKI FEEDEISHLV P |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
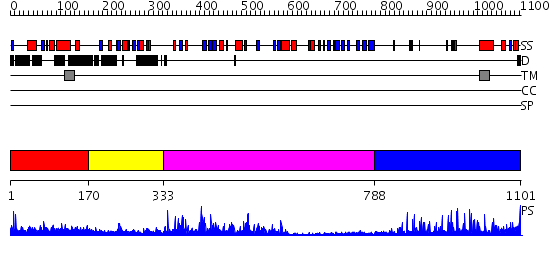
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..169] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [170..332] | 13.522879 | High Resolution Crystal Structure of the Intact Pro-Kumamolisin, a Sedolisin Type Proteinase (previously called Kumamolysin or KSCP) | |
| 3 | View Details | [333..787] | 93.69897 | Furin, C-terminal domain; Furin, N-terminal domain | |
| 4 | View Details | [788..1101] | 11.30103 | Low density lipoprotein (LDL) receptor YWTD domain; Low density lipoprotein (LDL) receptor, different EGF domains; Ligand-binding domain of low-density lipoprotein receptor |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|