













| General Information: |
|
| Name(s) found: |
CG5728-PA /
FBpp0084016
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1430 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC]
nucleolus [ISS] |
| Biological Process: |
regulation of alternative nuclear mRNA splicing, via spliceosome
[IMP]
rRNA processing [ISS] |
| Molecular Function: |
mRNA binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVPNEKSFPR GGTIHSEVKT DDVSLNIVFG ASQKKVKKAP KVKENLLSYE TEEQNGQLEA 60
61 FSAETLNMDT LQEDMLVMGV VKELTATALQ IALPGRMFAR TLVADISDAY TRVAKAAMSG 120
121 DTSEYHDLTE LFQLGRIVYG KAIKTEKLDT GRVSLLLSLK PADVHGNLHH KSIKKGFIFS 180
181 GAVAEALEHG YVIESGVQGL QAFVPCEKPA QKLHVGQLAF LKVKTVHHDT HQSTCTCVQV 240
241 EQDQLRIKSQ NETNLDYILP GSIVKFKVAK HLKDGLKGSI MNESFSAYVN EHHLANALDT 300
301 LDAYELNEDY NARVLYVMPL TKLVYLTLNL DIKTGAAVAK DQDEEEQEVE PIKVGSVVEK 360
361 AKVLRLGSGG VVLLLNKKLK GIISYGSIRG NFKGNYDKDE VLSKYGRKTK HKVRILGYDV 420
421 IESLYYCSDD PNVVNEKLFC LEDINAGDLV TAKIFKKDDK IKGWSVRIGK VNGILEQFYL 480
481 APNVRYDVGQ SLKCRVLEVN AERKICYVSN RAEYLGKGIK ILTDYASAHV GNVYMGTVVR 540
541 CEDTYVLVKF GNGIKGVLHR QNLKENSSFF EGQTTKFRIL TRNKDQITLT LPEDKFQLGE 600
601 ICPVEITNAL DAGLEVKITF AAEDDEEDED GNPKLEEFVG LIPLRLLSDH LELLHAQMRV 660
661 HPAGSYTDAA CIMQNIFSLR DVPYFSGQLT KDWQSVQVGD IIRSYVKHAT DQVVDLMVCV 720
721 RNYNKPVKVH VKMLRLNAVK NAPVELVPEQ LLWVKVLSKE VETKTLTVSA KLTDVWSGDL 780
781 SDTAKLVEGY LNEVAQIKAG LEEASAPISK YSVGEKINVV FKGIDATTND WVYTVEGNGK 840
841 VSALLLSSLV GTAKAPEMGS KHEAVILWIE YSSDVLLISN KKLDIAHISP SGELSTNLIG 900
901 KAGMKAKVLL KLESVAVCSL KKGTNPLVIC PIRLHPNDIE NSGSAELRQG DFCNIAFIHD 960
961 KLHIAVPETV WRLWRGVKRT AGTEVAPVKA KKAKVEESPK QKKTTIEETS AQKKVKAKAK1020
1021 VETKTEAKAI PTKRKAEVVE KITNGQKKTQ PLTNGIVKEK PKQNGKLFFE DKTPAKNAKS1080
1081 ETPKSNGAEG KSRLPGVSSF WEDDVNQSKE GSSDEDEELN VAETQKNAAK KKRLSAKEKA1140
1141 KAEIKEEQRL REIEERNADP KARLETIDQY ERLVIAQPNN SISWLKYIAF LLSNTEIEKA1200
1201 RALARRAIST ISFRETQELR NMWSALLNME LVYSNNFDDV LKEALNCNDP LEIYISVVDI1260
1261 LKKNKRKDRL SSVLTTVLNK FKTELRVWPV AAEAYFWLGK SDQVHNLLQR ALRALPNQEH1320
1321 IPCIVSFAKL YAKHDNNDMA QTLLDDVVTS YPKRIDIWSV YVDMLIKAGL IDSARNVLER1380
1381 AVVQKLKPNK MQVIYKKYLQ LEENHGTDAT VAKVKQQAEQ WVKNYAKTVK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
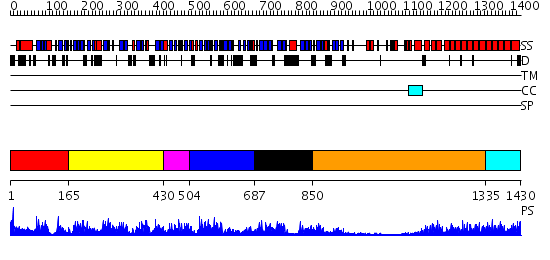
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..164] | 3.68 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 2 | View Details | [165..429] | 16.154902 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 3 | View Details | [430..503] | 25.30103 | No description for 2oceA was found. | |
| 4 | View Details | [504..686] | 6.522879 | Archaeal exosome core | |
| 5 | View Details | [687..849] | 4.69897 | C-terminal domain of RNA polymerase II subunit RBP4 (RpoE); N-terminal, heterodimerisation domain of RBP4 (RpoE) | |
| 6 | View Details | [850..1334] | 8.39794 | No description for 2ooeA was found. | |
| 7 | View Details | [1335..1430] | 64.09691 | The superhelical TPR domain of O-linked GlcNAc transferase reveals structural similarities to importin alpha. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)