













| General Information: |
|
| Name(s) found: |
DNApol-epsilon-PA /
FBpp0083800
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2236 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
epsilon DNA polymerase complex [ISS] |
| Biological Process: |
DNA replication
[IEA]
DNA-dependent DNA replication [IDA] response to DNA damage stimulus [IMP] nucleobase, nucleoside, nucleotide and nucleic acid metabolic process [IEA] |
| Molecular Function: |
nucleotide binding
[IEA]
DNA binding [IEA] DNA-directed DNA polymerase activity [IDA][ISS][NAS] zinc ion binding [IEA] 3'-5'-exodeoxyribonuclease activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDSSKGKVL QNTGKFVSEN RTEGDDFFNE AGYRQSREND KIDSKYGFDR VKDSQERTGY 60
61 LINMHSNEVL DEDRRLIAAL DLFFIQMDGS RFKCTVAYQP YLLIRPEDNM HLEVARFLGR 120
121 KYSGQISGLE HITKEDLDLP NHLSGLQQQY IKLSFLNQTA MTKVRRELMS AVKRNQERQK 180
181 SNTYYMQMLA TSLAQSSAGS EDATLGKRQQ DYMDCIVDIR EHDVPYHVRV SIDLRIFCGQ 240
241 WYNIRCRSGV ELPTITCRPD ILDRPEPVVL AFDIETTKLP LKFPDAQTDQ VMMISYMIDG 300
301 QGYLITNREI ISSNVDDFEY TPKPEFEGNF IVFNEENEMQ LLQRFFDHIM EVRPHIIVTY 360
361 NGDFFDWPFV ETRAAVYDLD MKQEIGFSKL RDGNYLSRPA IHMDCLCWVK RDSYLPVGSQ 420
421 GLKAVAKAKL RYDPVELDPE DMCRMAVEQP QVLANYSVSD AVATYYLYMK YVHPFIFALN 480
481 TIIPMEPDEI LRKGSGTLCE TLLMVEAYHA QIVYPNKHQS ELNKLSNEGH VLDSETYVGG 540
541 HVEALESGVF RADIPCRFRL DPAMVKQLQE QVDAVLRHAI EVEEGIPLEK VLNLDEVRQE 600
601 IVQGLQGLHD IPNRLEQPVI YHLDVGAMYP NIILTNRLQP SAMVSDLDCA ACDFNKPGVR 660
661 CKRSMDWLWR GEMLPASRNE FQRIQQQLET EKFPPLFPGG PQRAFHELSK EDQAAYEKKR 720
721 LTDYCRKAYK KTKLTKLETR TSTICQKENS FYVDTVRAFR DRRYEYKGLT KVAKASVNAA 780
781 VASGDAAEIK AAKGREVLYD SLQLAHKCIL NSFYGYVMRR GARWHSMPMA GIVCLTGSNI 840
841 ITKAREIIER VGRPLELDTD GIWCILPGSF PQEFTIHTSH EKKKKINISY PNAVLNTMVK 900
901 DHFTNDQYHE LRKDKENNLP KYDIRDENSI FFEVDGPYLA MVLPAAKEEG KKLKKRYAVF 960
961 NFDGTLAELK GFEVKRRGEL QLIKNFQSSV FEAFLAGSTL EECYASVAKV ADYWLDVLYS1020
1021 RGSNLPDSEL FELISENKSM SKKLEEYGAQ KSTSISTAKR LAEFLGEQMV KDAGLACKYI1080
1081 ISKKPEGAPV TERAIPLAIF QSEPSVRRHH LRRWLKDNTM GDADIRDVLD WNYYIERLGG1140
1141 TIQKIITIPA ALQGLANPVP RVQHPDWLHK KMLEKNDVLK QRRINEMFTS RPKPKPLATE1200
1201 EDKLADMEDL AGKDGGEGAA GCPIVTKRKR IQLEEHDDEE AQPQATTWRQ ALGAPPPIGE1260
1261 TRKTIVEWVR FQKKKWKWQQ DQRQRNRQAS KRTRGEDPPV VRATGSTATL GGFLRRAQRT1320
1321 LLDQPWQIVQ LVPVDDLGHF TVWALIGEEL HKIKLTVPRI FYVNQRSAAP PEEGQLWRKV1380
1381 NRVLPRSRPV FNLYRYSVPE QLFRDNSLGM LADLATPDIE GIYETQMTLE FRALMDMGCI1440
1441 CGVQREEARR LAQLATKDLE TFSIEQLEQR PQTQVKYLAS ANNRLRKIYL YQHNTPTAKK1500
1501 EIWSLILMPS KKAFVFALDT VRANQMPNMR QLYTAERLAL LKNLTAEEQD KIPVEDYTFE1560
1561 VLIEVDVKQI YRHIQRALTT YKQEHQGPTI LCLQTALSAR KLSLAMPILL EFPQAEIHIS1620
1621 DDASLLSGLD WQRQGSRAVI RHFLNLNNVL DLMLDQCRYF HVPIGNMPPD TVLFGADLFF1680
1681 ARLLQRHNFV LWWSASTRPD LGGREADDSR LLAEFEESIS VVQNKAGFYP DVCVELALDS1740
1741 LAVSALLQST RIQEMEGASS AITFDVMPQV SLEEMIGTVP AATLPSYDET ALCSAAFRVM1800
1801 RSMVNGWLRE VSINRNIFSD FQIVHFYRWV RSSNALLYDP ALRRSLNNLM RKMFLRIIAE1860
1861 FKRLGATIIY ADFNRIILSS GKKTVSDALG YVDYIVQSLR NKEMFHSIQL SFEQCWNFML1920
1921 WMDQANFSGI RGKLPKGIDE TVSSIVSTTM IRDSERNQDD DEDEEEDSEN RDPVESNEAE1980
1981 QDQEDELSLE LNWTIGEHLP DENECREKFE SLLTLFMQSL AEKKTTEQAI KDISHCAFDF2040
2041 ILKLHKNYGK GKPSPGLELI RTLIKALSVD KTLAEQINEL RRNMLRLVGI GEFSDLAEWE2100
2101 DPCDSHIINE VICKACNHCR DLDLCKDKHR AMKDGVPVWL CAQCYVAYDN EEIEMRMLDA2160
2161 LQRKMMSYVL QDLRCSRCSE IKRENLAEFC TCAGNFVPLI SGKDIQTLLG TFNKVAANHK2220
2221 MQLLQQTVHQ ALTTPR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
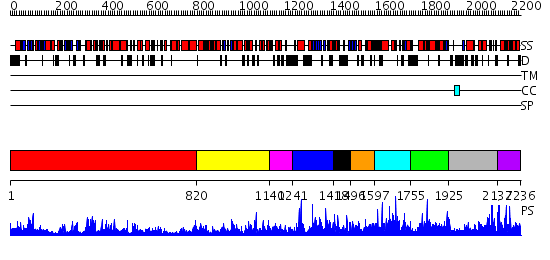
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..819] | 98.69897 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 2 | View Details | [820..1139] | 4.18 | Crystal structure of ESCHERICHIA coli DNA Polymerase II | |
| 3 | View Details | [1140..1240] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [1241..1417] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1418..1495] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1496..1596] | 1.040979 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1597..1754] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1755..1924] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1925..2136] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [2137..2236] | 9.050987 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)