













| General Information: |
|
| Name(s) found: |
FBpp0311985
[FlyBase]
E2f-PA / FBpp0083518 [FlyBase] E2f-PC / FBpp0083517 [FlyBase] E2f-PB / FBpp0083516 [FlyBase] |
| Description(s) found:
Found 58 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 805 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][NAS]
Rb-E2F complex [IDA] |
| Biological Process: |
positive regulation of cell proliferation
[NAS]
DNA endoreduplication [IGI][IMP] imaginal disc development [TAS] positive regulation of transcription [TAS] regulation of transcription, DNA-dependent [IEA] muscle tissue development [IMP] positive regulation of nurse cell apoptosis [IMP] nurse cell apoptosis [TAS] neuron development [IMP] growth [NAS] cellular process [IMP] dendrite morphogenesis [IMP] eggshell chorion gene amplification [TAS] regulation of transcription involved in G1/S-phase of mitotic cell cycle [NAS] positive regulation of gene expression [IMP] G1/S transition of mitotic cell cycle [TAS] regulation of cell cycle [IMP][NAS] antimicrobial humoral response [IMP] |
| Molecular Function: |
DNA binding
[IDA]
transcription factor activity [IEA] protein binding [IPI] RNA polymerase II transcription factor activity [IDA][NAS] transcription activator activity [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKFFVNVAP INNSNSSSSH TTTSSNTQRH QQHQQHYGGS GTTGHTMVAR RLNYDLHGGT 60
61 TSINNNNNIV IKNESVDLDY DHVLSSSDSN SNGGVAAHLR DHVYISLDKG HNTGAVATAA 120
121 AAATAGHTQQ QLQQQHHHQN QQQRKATGKS NDITNYYKVK RRPHAVSDEI HPKKQAKQSA 180
181 HHQTVYQKHT ASSAPQQLRH SHHQLRHDAD AELDEDVVER VAKPASHHPF SLSTPQQQLA 240
241 ASVASSSSSG DRNRADTSLG ILTKKFVDLL QESPDGVVDL NEASNRLHVQ KRRIYDITNV 300
301 LEGINILEKK SKNNIQWRCG QSMVSQERSR HIEADSLRLE QQENELNKAI DLMRENLAEI 360
361 SQEVENSGGM AYVTQNDLLN VDLFKDQIVI VIKAPPEAKL VLPNTKLPRE IYVKAENSGE 420
421 INVFLCHDTS PENSPIAPGA GYVGAPGAGC VRTATSTRLH PLTNQRLNDP LFNNIDAMST 480
481 KGLFQTPYRS ARNLSKSIEE AAKQSQPEYN NICDIAMGQH HNLNQQQQQQ QQQLLQQPEE 540
541 DDVDVELNQL VPTLTNPVVR THQFQQHQQP SIQELFSSLT ESSPPTPTKR RREAAAAAIA 600
601 AGSSTTATTT LNSHNNRNHS NHSNHSNHSS SNNSKSQPPT IGYGSSQRRS DVPMYNCAME 660
661 GATTTSATAD TTAATSRSAA ASSLQMQFAA VAESNNGSSS GGGGGGGGYG SIAGAGANAD 720
721 PHQPYSHDRN SLPPGVADCD ANSNSSSVTL QGLDALFNDI GSDYFSNDIA FVSINPPDDN 780
781 DYPYALNANE GIDRLFDFGS DAYGP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
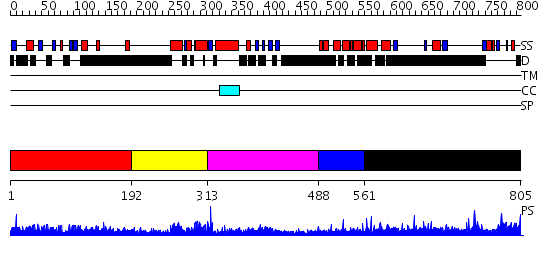
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..191] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [192..312] | 26.0 | Cell cycle transcription factor E2F-4 | |
| 3 | View Details | [313..487] | 22.69897 | Structure of the Rb C-terminal domain bound to an E2F1-DP1 heterodimer | |
| 4 | View Details | [488..560] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [561..805] | 5.129996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)