













| General Information: |
|
| Name(s) found: |
Abd-B-PB /
FBpp0082826
[FlyBase]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 493 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][NAS]
|
| Biological Process: |
negative regulation of salivary gland boundary specification
[TAS]
midgut development [TAS] male gonad development [IMP] external genitalia morphogenesis [IMP] genital disc sexually dimorphic development [IMP] salivary gland development [NAS] regulation of transcription [IEA] negative regulation of striated muscle tissue development [IMP] negative regulation of cardioblast cell fate specification [IMP] gonadal mesoderm development [IMP] developmental pigmentation [NAS] heart development [NAS] positive regulation of developmental pigmentation [NAS] sex differentiation [NAS] female genitalia development [IMP] genital disc development [TAS] spiracle morphogenesis, open tracheal system [IMP] pole cell migration [TAS] anterior/posterior axis specification, embryo [NAS] specification of segmental identity, abdomen [IMP][NAS] imaginal disc-derived genitalia development [TAS] regulation of transcription, DNA-dependent [IEA] germ cell migration [IMP][TAS] imaginal disc-derived female genitalia development [IMP] open tracheal system development [IMP] gonad development [NAS] male genitalia development [IMP] genital disc anterior/posterior pattern formation [IEP] determination of genital disc primordium [IMP] sex-specific pigmentation [TAS] |
| Molecular Function: |
transcription factor activity
[IDA]
specific RNA polymerase II transcription factor activity [NAS] sequence-specific DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQQHHLQQQQ QQQQQQEQQH LQEQQQHLQQ LHHHAHHHLP QPLHTTSHHH SAHPHLQQQQ 60
61 QQQQHAVVAS SPSSVLQQQQ QQSTPTTHST PTHAVMYEDP PPVPLVAVQQ QHLPAPQQQQ 120
121 QLQQQQQQQQ QQLATTPVAG ALSPAQTPTG PSAQQQQHLT SPHHQQLPQQ QTPNSVASGA 180
181 SSNLQQQQQQ QNAAVAPGQT QIVAPTTASV SPSSVSSQKE DINMSIQLAP LHIPAIRAGP 240
241 GFETDTSAAV KRHTAHWAYN DEGFNQHYGS GYYDRKHMFA YPYPETQFPV GQYWGPNYRP 300
301 DQTTSAAAAA AYMNEAERHV SAAARQSVEG TSTSSYEPPT YSSPGGLRGY PSENYSSSGA 360
361 SGGLSVGAVG PCTPNPGLHE WTGQVSVRKK RKPYSKFQTL ELEKEFLFNA YVSKQKRWEL 420
421 ARNLQLTERQ VKIWFQNRRM KNKKNSQRQA NQQNNNNNSS SNHNHAQATQ QHHSGHHLNL 480
481 SLNMGHHAAK MHQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
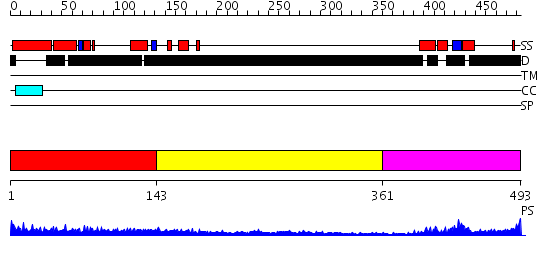
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..142] | 4.522879 | Sec24 | |
| 2 | View Details | [143..360] | 1.254995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [361..493] | 18.522879 | Crystal Structure of HoxA9 and Pbx1 homeodomains bound to DNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)