













| General Information: |
|
| Name(s) found: |
mor-PA /
FBpp0082692
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1209 amino acids |
Gene Ontology: |
|
| Cellular Component: |
brahma complex
[IDA][NAS][TAS]
chromatin remodeling complex [ISS] |
| Biological Process: |
chromatin remodeling
[NAS]
oogenesis [NAS] positive regulation of transcription, DNA-dependent [IDA] |
| Molecular Function: |
DNA binding
[IEA]
protein binding [IPI] transcription coactivator activity [IC] general RNA polymerase II transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNTLGPKKDG SPNIDFFQSP ETLQGFESIR QWLQKNCKKY LAHSSEPITK ESLAQLLIHF 60
61 LQYVEAKLGK NSADPPATRI PMRCFLDFKS GGGLCIIFST MFRFRAEQRG KKFDFSIGKN 120
121 PTRKDPNIQL LIEIEQALVE ADLYRIPYIY IRPEIEKGFE GKLREILDNR RVEIVSDEED 180
181 ATHIVYPVVD PHPDEYARPI FKRGGHVMLH WYYFPESYDS WAVNNFDLPD HIPENPESPA 240
241 ERWRVSASWI VDLEQYNEWM AEEDYEVDEQ GKKKTHKQRM SIDDIMSFGD EKKKPAASSG 300
301 GGKQKRRRSP SPASSASTSK PGKRKRSPAV VHKKSRNDDD DEDLTRDLDD PPAEPNVQEV 360
361 HKANAALQST ASPAPGGKSR GDNDMMPIKG GTMTDLDDEM TGGSAAQAMS TGDGENSQTG 420
421 KTSDNSNTQE FSSSAKEDME DNVTEQTHHI IVPSYSAWFD YNSIHVIEKR AMPEFFNSKN 480
481 KSKTPEIYMA YRNFMIDTYR LNPTEYLTST ACRRNLAGDV CAIMRVHAFL EQWGLINYQI 540
541 DADVRPTPMG PPPTSHFHIL SDTPSGLQSI NPQKTQQPSA AKTLLDLDKK PLGKDGGLEL 600
601 GDKSGLTGIK TEALENGAAG GLSSGVSQFG LKLDQYAKKP AAMRNRTAAS MAREWTDQET 660
661 LLLLEGLEMH KDDWNKVCEH VGSRTQDECI LHFLRLPIED PYLEDDGGFL GPLACQPIPF 720
721 SKSGNPIMST VAFLASVVDP RVAAAAAKAA MEEFAAIKDE VPATIMDNHL KNVEKASAGG 780
781 KFNPNFGLAN SGIAGTGNDK DDEEGKEGGA SASAGGSDEE MKDLSKKDDD AKSKDNTKSD 840
841 KTNTNTDSSS TSSSATGNTN NTDKKPKESS GSSPSGDKSA IKSDKSNKSS PTETAAAASG 900
901 GEVDIKTEDS SGDGETKDGT EAKEGTGTGT GPLAVPKEGT FSENNMQTAA AAALASAAVK 960
961 AKHLAALEER KIKSLVALLV ETQMKKLEIK LRHFEELEAT MEREREGLEY QRQQLITERQ1020
1021 QFHLEQLKAA EFRARQQAHH RLQQELQGQA AAGSMILPQQ QQLPQAQQPQ QQQQTLPPHP1080
1081 HLAQQQQLPP HPHQLPPQSQ PLAGPTAQHQ PLPPHVVSSP NGAPFAAPPI SLTGGLPPGA1140
1141 PTAIVTNPSD QTGPVAAGAA QSQAPTPMDT TPPSSGPVSD ANAPPGSAMP PGGIPPANTA1200
1201 PIAGVAPSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
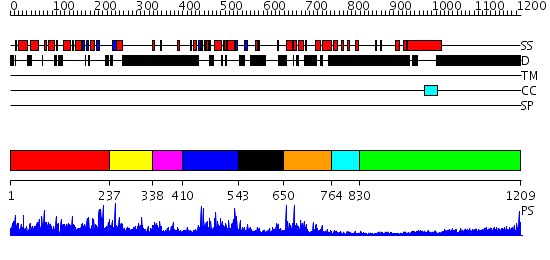
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..236] | 2.034931 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [237..337] | 11.522879 | Solution structure of the SANT domain of human KIAA1915 protein | |
| 3 | View Details | [338..409] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [410..542] | 39.0 | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes | |
| 5 | View Details | [543..649] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [650..763] | 5.522879 | No description for 2elkA was found. | |
| 7 | View Details | [764..829] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [830..1209] | 11.522879 | No description for 1y0fB was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)