













| General Information: |
|
| Name(s) found: |
FBpp0306662
[FlyBase]
rec-PA / FBpp0082647 [FlyBase] |
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 885 amino acids |
Gene Ontology: |
|
| Cellular Component: |
replication fork
[ISS]
pre-replicative complex [ISS] |
| Biological Process: |
reciprocal meiotic recombination
[IMP][NAS]
DNA replication [IEA][ISS] |
| Molecular Function: |
ATP binding
[IEA]
DNA binding [IEA] ATP-dependent DNA helicase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNPTLSRGGA PSRFVRGRGR GGGAYRPYFY FRRNGRVIPA GGNRQPNQGE PGAPDAPSVP 60
61 PATRQPRGWS RTAGKRGRNA HLDCSFLRPE NYAAPEDGLQ VQSIAVDNPR AYSGWRLYFL 120
121 REKYEEGNEL ATRIMAVEAY YQRNPHTFDF VMIRDRGFFP LCAAGIKSDA QLKEVWPSLS 180
181 EDIREHPLRT LGTLSLAMHT VVVNHQLDCN DSTTAMVSTP EQYVLPTPRV RKIYARPDDF 240
241 VPVESIEGIS HSRVDTLFSI RGFVSNVGEP SYSLTWQAFR CSRCQMEIAM RQRGTFQPRP 300
301 YQCKRSECVA RDEFVPLRSS PYTRLSIRQI IRVEESSLNL VHDFETSMPA EMDVELRHDL 360
361 VDAVRVGQEV VVTGILKLQE LGDDTTTGDT SNQMQPYLKA VSIRDASSIK REFSERDLEA 420
421 IVMINAEPNS FKLLVQSIAP EVYGHELPKA ACLLSLLGGK GAETEAINVL LVGDPGIGKT 480
481 KILQSCAQIA ERGAHVSGKR GAQSAQQLGV TFAGRNKRVL QAGSLMMASG GGHCTLDDVD 540
541 KLASKQAVLL QCLQSEEVNL PLAGAFASFP AQPSVIACAN PQRGQYDEGR YLLQNINISP 600
601 SLLREFHLVY ILLDKPSERD MSLTAHVRAL HAGARKRARI AARYALKPKM SDSMCEVSLN 660
661 VPAAGKDDDT IKTEDDNDSI MQQDYDLDKR LEVLPEEGDL DLLPPILIKK FLSYARQELN 720
721 PVLNEDASNA VLRYFLELKG SCNLDEDVSS QIGAGQLLAI IHLSQARARL DLSHVVSPQH 780
781 VRDVIALLTE SITQTSLKEG SSRQGTRGGG GAGGGAGKSA QLRNFLELTK RRSAALGRRI 840
841 FEFDELKEIG TRAGILTGFS QLVEMANLGG YLLMKGANMY EVVPD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
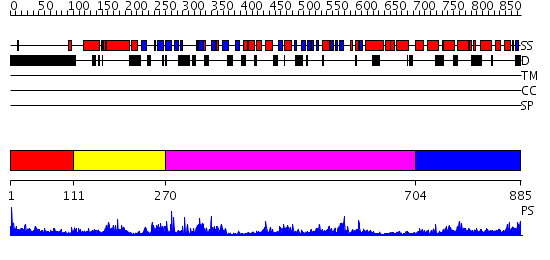
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..110] | 4.117997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [111..269] | 40.39794 | DNA replication initiator (cdc21/cdc54) N-terminal domain | |
| 3 | View Details | [270..703] | 14.0 | N-terminal, ClpS-binding domain of ClpA, an Hsp100 chaperone; ClpA, an Hsp100 chaperone, AAA+ modules | |
| 4 | View Details | [704..885] | 57.30103 | EDTA treated |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)