













| General Information: |
|
| Name(s) found: |
CG6194-PA /
FBpp0082569
[FlyBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 668 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
autophagy
[ISS]
autophagic cell death [IEP] proteolysis [ISS] salivary gland cell autophagic cell death [IEP] |
| Molecular Function: |
cysteine-type endopeptidase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNYDGLLSKL TDEKLMLFEA AGEGTIVEGS RICGQDADPL SLPLVEDGAI EEEQAAPIQT 60
61 IHRTVFAVPR VPSPSPVSTS GANLNLKTEN AATATPQRKI STSSRFMNAF SQLYGGGNSG 120
121 PSCGHVEQHN EEPGDLSANR TPTKGMESKL VAMWHNVKYG WSGKMRQTSF SKEQPVWLLG 180
181 RCYHRRFTPP VSMESSITEL PSGADTTPDN ATSAFDSIQA TSTSTSLYPA LNPQQIDEIV 240
241 VPQELGMDAV ENQVGEQPWE EGIEGFRRDF YSRIWMTYRR EFPIMNGSNY TSDCGWGCML 300
301 RSGQMLFAQG LICHFLGRSW RYDSESQLHS TYEDNMHKKI VKWFGDSSSK SSPFSIHALV 360
361 RLGEHLGKKP GDWYGPASVS YLLKHALEHA AQENADFDNI SVYVAKDCTI YLQDIEDQCS 420
421 IPEPAPKPHV PWQQAKRPQA ETTKTEQQQH WKSLIVLIPL RLGSDKLNPV YAHCLKLLLS 480
481 TEHCLGILGG KPKHSLYFVG FQEDRLIHLD PHYCQEMVDV NQENFSLHSF HCKSPRKLKA 540
541 SKMDPSCCIG FYCATKSDFD NFMESVQLYL HPMRCASGAT VDKAAGSHHT TPQSSHQTEQ 600
601 TEMNYPLFSF SRGRCMDHER DEMSDSLYKP LIKQVASLAQ ELGAQLAPPQ HGDEHDQDDS 660
661 ESEEFVLL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
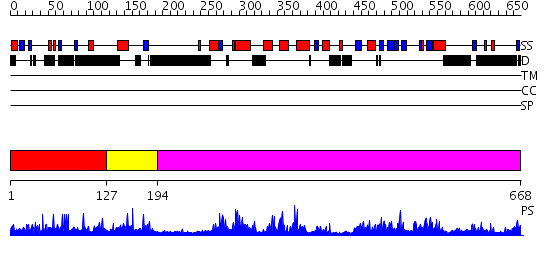
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..126] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [127..193] | 1.034987 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [194..668] | 147.0 | Structure of human Atg4b |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)