













| General Information: |
|
| Name(s) found: |
gi|74868822
[NCBI NR]
gi|24646843 [NCBI NR] gi|23171266 [NCBI NR] |
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1131 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apical plasma membrane
[IDA]
plasma membrane [IDA] |
| Biological Process: |
signal transduction
[IEA]
negative regulation of nucleobase, nucleoside, nucleotide and nucleic acid transport [IMP] |
| Molecular Function: |
3',5'-cyclic-GMP phosphodiesterase activity
[IDA][ISS]
3',5'-cyclic-nucleotide phosphodiesterase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHGPVSRSSS SSNMTDVSSP AGGAASPVEM STTSSSSAAT TSASSSKPLT NGANKTAIST 60
61 AAGGVTPGAV PGPGSGAIPA SSSSGNQVKL EHHHRQSNNN RPAVTNRSSE TKLMTPTGSS 120
121 SSPSQSPSQT QASIQTQTSQ QDRLAKASTT ASQQDVDEVA RLFEEKPEAF EKWLTERAPP 180
181 EALSRLQEFI ENRKPHKRPS VTSDLFQQWM AASPTVQQKS PRSLSNSSAS SLPECRRHLM 240
241 DLDEGELFME LIRDVANELD IDVLCHKILV NVGLLTHADR GSLFLAKGTP TNKYLVAKLF 300
301 DVTQKTALKD AVTRASAEEI IIPFGIGIAG MVAQTKQMIN IKEAYKDARF NCEIDLKTGY 360
361 KTNAILCMPI CNYEGDIIGV AQIINKTNGC MEFDEHDVEI FRRYLTFCGI GIQNAQLFEM 420
421 SVQEYRRNQI LLNLARSIFE EQNNLECLVT KIMTEARELL KCERCSVFLV DLDCCEASHL 480
481 EKIIEKPNQP ATRAIKSADS FEEKKMRNRF TVLFELGGEY QAANVSRPSV SELSSSTLAQ 540
541 IAQFVATTGQ TVNICDVIEW VRDHNQIRAE DEIDSTQAIL CMPIMNAQKK VIGVAQLINK 600
601 ANGVPFTDSD ASIFEAFAIF CGLGIHNTQM YENACKLMAK QKVALECLSY HATASQDQTE 660
661 KLTQDVIAEA ESYNLYSFTF TDFELVDDDT CRAVLRMFMQ CNLVSQFQIP YDVLCRWVLS 720
721 VRKNYRPVKY HNWRHALNVA QTMFAMLKTG KMERFMTDLE ILGLLVACLC HDLDHRGTNN 780
781 AFQTKTESPL AILYTTSTME HHHFDQCVMI LNSEGNNIFQ ALSPEDYRSV MKTVESAILS 840
841 TDLAMYFKKR NAFLELVENG EFDWQGEEKK DLLCGMMMTA CDVSAIAKPW EVQHKVAKLV 900
901 ADEFFDQGDL EKLQLNTQPV AMMDRERKDE LPKMQVGFID VICLPLYRVL CDTFPWITPL 960
961 YEGTLENRRN WQDLAEKVEM GLTWIDHDTI DKPVEEFAAC ADEEIKDIEF TVTTLNCNQQ1020
1021 SQHGSEDSHT PEHQRSGSRL SMKKTGALGK AVRSKLSKTL YNSMDGSKPK TSLKLLESHV1080
1081 SEDMDDKSPT SPSQPQASGS MGRMSASSST SSAGGQMVDK SKKRSKLCAL L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
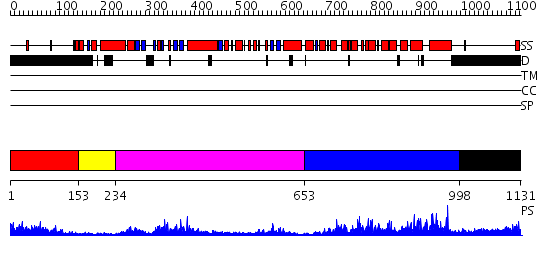
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..152] | 3.221849 | No description for 1y0fA was found. | |
| 2 | View Details | [153..233] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [234..652] | 51.221849 | 3',5'-cyclic nucleotide phosphodiesterase 2A, GAF A and GAF B domains | |
| 4 | View Details | [653..997] | 102.0 | Catalytic Domain Of Human Phosphodiesterase 5A In Complex With Tadalafil | |
| 5 | View Details | [998..1131] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)