













| General Information: |
|
| Name(s) found: |
svp-PB /
FBpp0082036
[FlyBase]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 543 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS][NAS]
cytoplasm [IDA] |
| Biological Process: |
nerve-nerve synaptic transmission
[IMP]
R1/R6 cell fate commitment [TAS] R7 cell fate commitment [TAS] cardioblast cell fate determination [IEP][IGI] phototaxis [IMP] regulation of transcription, DNA-dependent [IEA] R3/R4 cell fate commitment [TAS] regulation of transcription [ISS] fat body development [TAS] compound eye development [NAS] compound eye photoreceptor fate commitment [TAS] heart development [NAS][TAS] ventral cord development [TAS] central nervous system development [NAS] embryonic development via the syncytial blastoderm [NAS] |
| Molecular Function: |
transcription factor activity
[ISS]
zinc ion binding [IEA] protein binding [IPI] ligand-dependent nuclear receptor activity [ISS][NAS] steroid hormone receptor activity [IEA] sequence-specific DNA binding [IEA] receptor activity [NAS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCASPSTAPG FFNPRPQSGA ELSAFDIGLS RSMGLGVPPH SAWHEPPASL GGHLHAASAG 60
61 PGTTTGSVAT GGGGTTPSSV ASQQSAVIKQ DLSCPSLNQA GSGHHPGIKE DLSSSLPSAN 120
121 GGSAGGHHSG SGSGSGSGVN PGHGSDMLPL IKGHGQDMLT SIKGQPTGCG STTPSSQANS 180
181 SHSQSSNSGS QIDSKQNIEC VVCGDKSSGK HYGQFTCEGC KSFFKRSVRR NLTYSCRGSR 240
241 NCPIDQHHRN QCQYCRLKKC LKMGMRREAV QRGRVPPTQP GLAGMHGQYQ IANGDPMGIA 300
301 GFNGHSYLSS YISLLLRAEP YPTSRYGQCM QPNNIMGIDN ICELAARLLF SAVEWAKNIP 360
361 FFPELQVTDQ VALLRLVWSE LFVLNASQCS MPLHVAPLLA AAGLHASPMA ADRVVAFMDH 420
421 IRIFQEQVEK LKALHVDSAE YSCLKAIVLF TTDACGLSDV THIESLQEKS QCALEEYCRT 480
481 QYPNQPTRFG KLLLRLPSLR TVSSQVIEQL FFVRLVGKTP IETLIRDMLL SGNSFSWPYL 540
541 PSM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
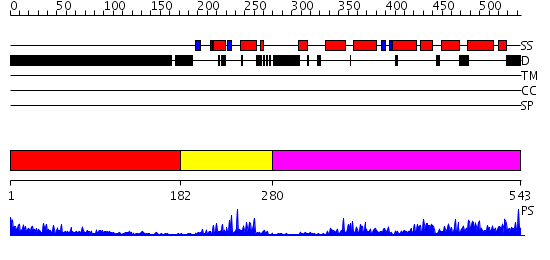
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..181] | 17.237996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [182..279] | 20.0 | Crystal Structure of Vitmain D Receptor and 9-cis Retinoic Acid Receptor DNA-Binding Domains Bound to a DR3 Response Element | |
| 3 | View Details | [280..543] | 65.69897 | LIGAND-BINDING DOMAIN OF THE HUMAN NUCLEAR RECEPTOR RXR-ALPHA |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)