













| General Information: |
|
| Name(s) found: |
ftz-PA /
FBpp0081139
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 410 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC][IEA][NAS]
|
| Biological Process: |
posterior head segmentation
[TAS]
segmentation [IMP] blastoderm segmentation [TAS] periodic partitioning by pair rule gene [NAS][TAS] regulation of transcription, DNA-dependent [IEA][ISS] trunk segmentation [TAS] germ cell migration [IMP] regulation of transcription [IEA][IMP] regulation of transcription from RNA polymerase II promoter [NAS] gonadal mesoderm development [IMP] central nervous system development [TAS] negative regulation of transcription, DNA-dependent [IDA] cell fate specification [TAS] |
| Molecular Function: |
transcription factor activity
[ISS]
sequence-specific DNA binding [IDA] transcription repressor activity [IDA][IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATTNSQSHY SYADNMNMYN MYHPHSLPPT YYDNSGSNAY YQNTSNYQGY YPQESYSESC 60
61 YYYNNQEQVT TQTVPPVQPT TPPPKATKRK AEDDAASIIA AVEERPSTLR ALLTNPVKKL 120
121 KYTPDYFYTT VEQVKKAPAV STKVTASPAP SYDQEYVTVP TPSASEDVDY LDVYSPQSQT 180
181 QKLKNGDFAT PPPTTPTSLP PLEGISTPPQ SPGEKSSSAV SQEINHRIVT APNGAGDFNW 240
241 SHIEETLASD CKDSKRTRQT YTRYQTLELE KEFHFNRYIT RRRRIDIANA LSLSERQIKI 300
301 WFQNRRMKSK KDRTLDSSPE HCGAGYTAML PPLEATSTAT TGAPSVPVPM YHHHQTTAAY 360
361 PAYSHSHSHG YGLLNDYPQQ QTHQQYDAYP QQYQHQCSYQ QHPQDLYHLS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
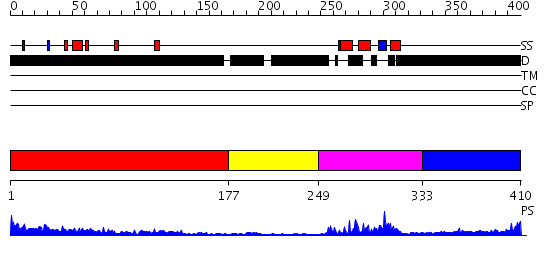
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..176] | 12.525996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [177..248] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [249..332] | 23.39794 | DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF THE ANTENNAPEDIA HOMEODOMAIN FROM DROSOPHILA IN SOLUTION BY 1H NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | |
| 4 | View Details | [333..410] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)