













| General Information: |
|
| Name(s) found: |
fab1-PB /
FBpp0088952
[FlyBase]
fab1-PA / FBpp0088523 [FlyBase] |
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1809 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endosome
[IDA]
vesicle membrane [ISS] |
| Biological Process: |
intracellular signaling pathway
[ISS]
cellular protein metabolic process [IEA] phosphoinositide phosphorylation [IMP] phosphatidylinositol metabolic process [IEA] endosome to lysosome transport [IMP] phosphorylation [NAS] |
| Molecular Function: |
ATP binding
[IEA]
zinc ion binding [ISS] 1-phosphatidylinositol-3-phosphate 5-kinase activity [IMP] protein binding [IEA] 1-phosphatidylinositol-4-phosphate 5-kinase activity [ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSNNQNNSS SHQHLHSPSK LTEFARNFED KPESLFGRVV NKIQNVYNQS YNTVNDISSG 60
61 SSSSSSTQPV QVVGKSQFFS DSQTSTAEIA DVETSSQSSV RPQPPTTLSI RTNSETRGTS 120
121 TSSNTAAEDS ETSDRVETLP LPTSEANQGR TVSNVLKHIS NIVATKNNND LRNYKDTELQ 180
181 RFWMPDSKAK ECYDCSQKFS TFRRKHHCRL CGQIFCSKCC NQVVPGMIIR CDGDLKVCNY 240
241 CSKIVLTFLK SSSSEMGQDM QELQQHLSNK LEVQDSGSSL AKHPQMQRAP LPRKTSVGYQ 300
301 EERFSSHPTY TTLSIDDRKN ILQQSNSLIT LHEEMQRDLP AQNCGQRLIE FLNSNNKSAN 360
361 EVQAVAILNA MLAAGFLEPI VPDPEQMDFD SSLHYKFSKS SSSDTSRTMS PQFEANPHAE 420
421 PQPPKSMDQS AEEKEKELEN ELENDRCYTT ATSKLLASYC EHEEQLLAQM LRAHNLDQEW 480
481 DKVLQMLCST AANHFKPEHC SNDLMDIRNY VNFKKVPGGR RKDSKIVHGV AFSKNVAHKD 540
541 MATHVPFPRI LLLQCPIVYE RIEGKFVTIE TVLLQEKEYL RNVCARIMSF KPNVVLVHKN 600
601 VAGIAQDLLR SYEVTLVLDV KLSVMERLSR TLQCDIVSSI ESNITMPKLG YCNDFYIRNY 660
661 NGKTLMFFEK LTNPRGYTCL LRGGSNAELT RVKRVASALL FARYNWRLEM SFLLNEFAQP 720
721 LSPKPSIFDS KETSPKTETE AELRSKRPII LERKSEDKIT TIVSENVSDF TDPLRASQAE 780
781 ALSTSPCAPP VVEALAVEPR YDNRFRTALS STLLSVSPFL TFPLPYLETE QGRKCKLRKL 840
841 FPAELYFSKQ WSRTGLERPD SMGDGEAGKS EPGNKENQMQ LLPAHDFVLM KITAPASSRD 900
901 IQSKLAEFRS FGGRLPKGKA PMLRPKKKNA EVIQRPQKVS EEQLYKDALD PQNHQRLPVL 960
961 FCSFHYNPKG VSSFCKLPML LDMKFYGQYD IMLEQFLQRY CCLFNSMCPS CNLPMLGHVR1020
1021 RYVHSLGCVH VYLTEDLTRS DPTRIYFTSW CSICNATTPT IPLSDAAKCL SLAKYLEMRF1080
1081 HGHAYKRRPP STDAEQGGTV CEHSLHRDYV HHFSFRGVGA KFQYTPVEVW ETDLPSLTVQ1140
1141 LDLPQPFQSA QVQEEIKNFS IKGHEVYNRI HERIADLATE EENSPLVQHL KTMLTHDQFI1200
1201 FKQKIEIVHT LLTDNRATAY DTSDALAMAR RALAESIELW GPRLQEIEKL TAKQAHHIDS1260
1261 GTICTEELRP EQVQTADSSK VTTSSLPKEN DPLECPSEDT ETGASNSQTV LDKNFSIDQM1320
1321 LASTVNVYSD KKSIRKILTQ LLPSGNQVNP LQSPFPAQDH LTLPLGSIPI HVRETDLSSV1380
1381 IAYSLTSMDY QKAIDEAEAN SNAAHSSPQL KRKIPLAESV SDAEDSPSLS RTSSNTSAAP1440
1441 NASVPSPATA ASESEEKSKE RIKQPPSPHI TLAFQDHSCQ FQCKIYFARE FDAMRSKSLK1500
1501 PPKLDKSLYR RLEKSKMREE LRISQSRTGS EMELVRKPSD VGAPRTTEDD SNQEEDARIA1560
1561 LARSLCKSVQ WEARGGKSGS RFCKTLDDRF VLKEMNSRDM TIFEPFAPKY FEYIDRCQQQ1620
1621 QQPTLLAKIF GVFRVSVKKK DSFVERSVMV MENLFYGCNI ENKFDLKGSE RNRLVDPSNQ1680
1681 QGEIVLLDEN LVQMSWSKPL YVLSHSKTVL RDAIQRDSSF LEKNLVMDYS LLVGLDKKNG1740
1741 VLVLGIIDYI RTFTLDKRVE SIIKGSGILG GKGKDPTVVN PERYKQRFID AMDRYFLTVP1800
1801 DRWEGLSKV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
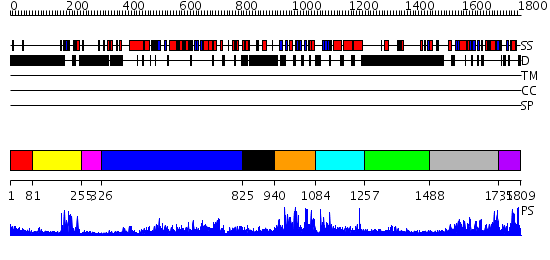
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [81..254] | 24.0 | Hrs | |
| 3 | View Details | [255..325] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [326..824] | 119.0 | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) | |
| 5 | View Details | [825..939] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [940..1083] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1084..1256] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1257..1487] | 5.127996 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1488..1734] | 63.30103 | Phosphatidylinositol phosphate kinase IIbeta, PIPK IIbeta | |
| 10 | View Details | [1735..1809] | 3.09 | No description for 2gk9A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)