













| General Information: |
|
| Name(s) found: |
FBpp0303577
[FlyBase]
FBpp0303575 [FlyBase] |
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1504 amino acids |
Gene Ontology: |
|
| Cellular Component: |
extracellular region
[IDA]
proteinaceous extracellular matrix [TAS] |
| Biological Process: |
negative chemotaxis
[IMP]
epithelial cell migration, open tracheal system [IMP] neuron migration [IMP] neuron differentiation [IMP] mesoderm migration [IMP] positive regulation of cell-cell adhesion [IMP] salivary gland boundary specification [IMP] axon guidance [IGI][IMP][IPI] axon midline choice point recognition [IMP] embryonic heart tube development [IMP] glial cell migration [IMP] |
| Molecular Function: |
protein binding
[IPI]
calcium ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAPSRTTLM PPPFRLQLRL LILPILLLLR HDAVHAEPYS GGFGSSAVSS GGLGSVGIHI 60
61 PGGGVGVITE ARCPRVCSCT GLNVDCSHRG LTSVPRKISA DVERLELQGN NLTVIYETDF 120
121 QRLTKLRMLQ LTDNQIHTIE RNSFQDLVSL ERLRLNNNRL KAIPENFVTS SASLLRLDIS 180
181 NNVITTVGRR VFKGAQSLRS LQLDNNQITC LDEHAFKGLV ELEILTLNNN NLTSLPHNIF 240
241 GGLGRLRALR LSDNPFACDC HLSWLSRFLR SATRLAPYTR CQSPSQLKGQ NVADLHDQEF 300
301 KCSGLTEHAP MECGAENSCP HPCRCADGIV DCREKSLTSV PVTLPDDTTE LRLEQNFITE 360
361 LPPKSFSSFR RLRRIDLSNN NISRIAHDAL SGLKQLTTLV LYGNKIKDLP SGVFKGLGSL 420
421 QLLLLNANEI SCIRKDAFRD LHSLSLLSLY DNNIQSLANG TFDAMKSIKT VHLAKNPFIC 480
481 DCNLRWLADY LHKNPIETSG ARCESPKRMH RRRIESLREE KFKCSWDELR MKLSGECRMD 540
541 SDCPAMCHCE GTTVDCTGRG LKEIPRDIPL HTTELLLNDN ELGRISSDGL FGRLPHLVKL 600
601 ELKRNQLTGI EPNAFEGASH IQELQLGENK IKEISNKMFL GLHQLKTLNL YDNQISCVMP 660
661 GSFEHLNSLT SLNLASNPFN CNCHLAWFAE WLRKKSLNGG AARCGAPSKV RDVQIKDLPH 720
721 SEFKCSSENS EGCLGDGYCP PSCTCTGTVV RCSRNQLKEI PRGIPAETSE LYLESNEIEQ 780
781 IHYERIRHLR SLTRLDLSNN QITILSNYTF ANLTKLSTLI ISYNKLQCLQ RHALSGLNNL 840
841 RVLSLHGNRI SMLPEGSFED LKSLTHIALG SNPLYCDCGL KWFSDWIKLD YVEPGIARCA 900
901 EPEQMKDKLI LSTPSSSFVC RGRVRNDILA KCNACFEQPC QNQAQCVALP QREYQCLCQP 960
961 GYHGKHCEFM IDACYGNPCR NNATCTVLEE GRFSCQCAPG YTGARCETNI DDCLGEIKCQ1020
1021 NNATCIDGVE SYKCECQPGF SGEFCDTKIQ FCSPEFNPCA NGAKCMDHFT HYSCDCQAGF1080
1081 HGTNCTDNID DCQNHMCQNG GTCVDGINDY QCRCPDDYTG KYCEGHNMIS MMYPQTSPCQ1140
1141 NHECKHGVCF QPNAQGSDYL CRCHPGYTGK WCEYLTSISF VHNNSFVELE PLRTRPEANV1200
1201 TIVFSSAEQN GILMYDGQDA HLAVELFNGR IRVSYDVGNH PVSTMYSFEM VADGKYHAVE1260
1261 LLAIKKNFTL RVDRGLARSI INEGSNDYLK LTTPMFLGGL PVDPAQQAYK NWQIRNLTSF1320
1321 KGCMKEVWIN HKLVDFGNAQ RQQKITPGCA LLEGEQQEEE DDEQDFMDET PHIKEEPVDP1380
1381 CLENKCRRGS RCVPNSNARD GYQCKCKHGQ RGRYCDQGEG STEPPTVTAA STCRKEQVRE1440
1441 YYTENDCRSR QPLKYAKCVG GCGNQCCAAK IVRRRKVRMV CSNNRKYIKN LDIVRKCGCT1500
1501 KKCY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
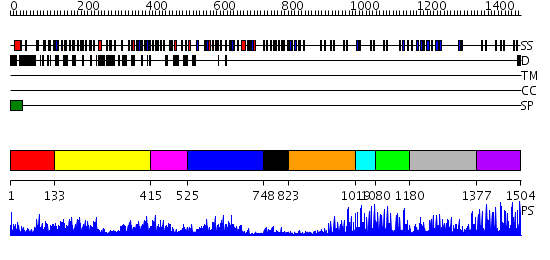
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..132] | 31.154902 | No description for 2v70A was found. | |
| 2 | View Details | [133..414] | 8.522879 | Ribonuclease inhibitor | |
| 3 | View Details | [415..524] | 35.154902 | No description for 2v9sA was found. | |
| 4 | View Details | [525..747] | 26.39794 | Third LRR domain of Drosophila Slit | |
| 5 | View Details | [748..822] | 4.34 | No description for 2o6rA was found. | |
| 6 | View Details | [823..1018] | 3.09691 | No description for 2dtgE was found. | |
| 7 | View Details | [1019..1079] | 10.69897 | No description for 2gy5A was found. | |
| 8 | View Details | [1080..1179] | 13.221849 | NMR structure of the human NOTCH-1 ligand binding region | |
| 9 | View Details | [1180..1376] | 5.77 | Modulation of agrin function by alternative splicing and Ca2+ binding | |
| 10 | View Details | [1377..1504] | 4.221849 | Coagulation factor VIIa |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|