













| General Information: |
|
| Name(s) found: |
cg-PA /
FBpp0086658
[FlyBase]
cg-PD / FBpp0086655 [FlyBase] |
| Description(s) found:
Found 39 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 671 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][NAS]
|
| Biological Process: |
imaginal disc-derived wing morphogenesis
[IMP]
|
| Molecular Function: |
nucleic acid binding
[IEA]
transcription factor activity [NAS] zinc ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCAAQNPPPF GYTWGFADNG NRAAESVLEI SPNINYTVSG ESMPYLLSTD GSLAVQKDVK 60
61 GLTAGGKNNV VRRMFVVNDP SFPPGTQRVI TAGGASTVVK KQDNNQQVLS LDKNCDILPG 120
121 ISVQVQKVIQ GLEDNEDSQG EAPNLKLEPG TLELSPKTEL QESMHFSETD ATIKKERPYS 180
181 CDECGKSFLL KHHLTTHARV HTGERPHICT HCGKSFAHKH CLNTHLLLHS TERPYQCQEC 240
241 KKSFTLKHHL LTHSRVHSRE RPFVCQECGR AFPLKRHLVT HSKFHAGERP YVCEECGESF 300
301 AQENHLIMHS RFHGSLNPFV CAECGASFPR KFQLVNHGRI HGKIPHSCTV CGKEFLQKRT 360
361 LVSHMRRVHT GEQAHPCVSC GEGFLTKAEL HQHVRTAHNG VNPNTSSATI IANQQLQQAH 420
421 HHQAGQQTHP QTITVVSNPG NSTLLTVSTT DANGVARPQF VCRECGSAFN SREALALHLR 480
481 LHTGDKSLMT DLCALTAALP GHFLSTASLN PGTVVTANPN LVGQSPVPVQ IISSTGQVMS 540
541 QTTLVQAANS THPQAVVTAV PTMPVHGQQQ HLQHVAQQQQ QQQQQQQQQQ QHVVSVAPAN 600
601 KPKSHFCASC GKGFAAKHGL MQHNRRHPNG GCTVRTHVCE CGKAFFQKNH LMLHQRQHLE 660
661 TKPAISQQQV C |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
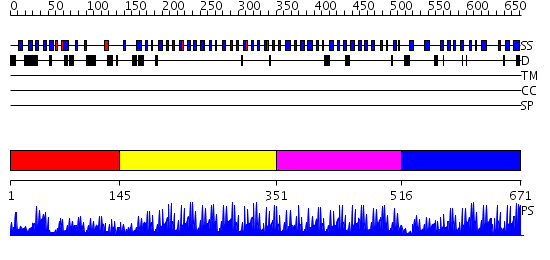
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..144] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [145..350] | 60.69897 | No description for 2i13A was found. | |
| 3 | View Details | [351..515] | 19.09691 | Five-finger GLI1 | |
| 4 | View Details | [516..671] | 5.045757 | THE CRYSTAL STRUCTURE OF A ZINC FINGER - RNA COMPLEX REVEALS TWO MODES OF MOLECULAR RECOGNITION |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)