













| General Information: |
|
| Name(s) found: |
Dp-PA /
FBpp0086853
[FlyBase]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 445 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[NAS]
Rb-E2F complex [IDA] Myb complex [IDA] |
| Biological Process: |
eggshell chorion gene amplification
[TAS]
oogenesis [IMP] positive regulation of gene expression [IMP] endomitotic cell cycle [TAS] regulation of transcription, DNA-dependent [IEA] dorsal/ventral axis specification, ovarian follicular epithelium [IMP] positive regulation of nurse cell apoptosis [IMP] nurse cell apoptosis [TAS] regulation of cell cycle [IMP][NAS] cell cycle [IMP] regulation of transcription from RNA polymerase II promoter [NAS] |
| Molecular Function: |
DNA binding
[IDA]
transcription factor activity [IEA] protein binding [IPI] RNA polymerase II transcription factor activity [NAS] specific RNA polymerase II transcription factor activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAHSTGGTVK TDEVNFFFRN EHGQISKMLK PAQNKSEMEG GKPVAVVYAT GSSARNSGSA 60
61 SGIGNVGRMG AFSQMGSQGQ FIRLQDNGLS IPKTEAGTTY TTVSAQKTSG AGSGHYDLPL 120
121 KGDRYVKFTP NPIKMKSKLH AIQSNSLHSM SASSSSVQRK RKPDKAGKGL RHFSMKVCEK 180
181 VEEKGKTTYN EVADDLVSEE MKNNAYDNNC DQKNIRRRVY DALNVLMAIN VISKDKKEIR 240
241 WIGLPANSTE TFLALEEENC QRRERIKQKN EMLREMIMQH VAFKGLVERN KRNESQGVVP 300
301 SPNASIQLPF IIVNTHKSTK INCSVTNDKS EYIFKFDKTF EMHDDIEVLK RMGFLLGLDK 360
361 GECTPENIER VKSWVPPNLA KYVEAYGTGK TGENMYESDD EDNEFNGYLE SANESQGFAQ 420
421 HSAQHTTDGE FKLEMDDDEL DDDID |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
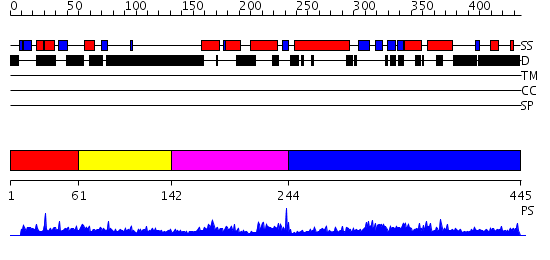
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..60] | 1.037977 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [61..141] | 2.118993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [142..243] | 30.30103 | Cell cycle transcription factor DP-2 | |
| 4 | View Details | [244..445] | 54.0 | Structure of the Rb C-terminal domain bound to an E2F1-DP1 heterodimer |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)