













| General Information: |
|
| Name(s) found: |
Sin3A-PD /
FBpp0087002
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1751 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS][NAS]
euchromatin [TAS] cytoplasm [IDA] Sin3 complex [NAS] chromatin [IDA] |
| Biological Process: |
muscle organ development
[IMP]
negative regulation of transcription, DNA-dependent [IMP][TAS] negative regulation of transcription [IEP] peripheral nervous system development [TAS] regulation of transcription, DNA-dependent [IEA] neuron development [IMP] regulation of transcription [ISS] regulation of mitotic cell cycle [IMP] dendrite morphogenesis [IMP] |
| Molecular Function: |
transcription factor activity
[ISS]
transcription cofactor activity [NAS] RNA polymerase II transcription factor activity [NAS] transcription repressor activity [IDA][IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMKRTRVDEV QFGTRPVPQT SGGVGVGVVG VTGGGPTSGG GGTATVGVNT TGVTIGTVVP 60
61 SAHNATISGI GSIHHRILTP QHGGAQTIAY LPSTTPTATN LKTTTSIVDS TTAGGPVGAG 120
121 AQVAVGVGSA AGGGGVVVST GSTGTQTLQY TTSYSVASIQ AGGTLKANTA DGANTVQIHV 180
181 TGGGAANNPA SAQTVSSSSQ TGTIRQRTIS GTQTVATAVG NLATISQQQP VQQSPLGKAQ 240
241 TPPSSVVANS IPVGGTTPPQ GQSGNATPRL KVEDALSYLD QVKYQYADQP QIYNNFLDIM 300
301 KEFKSHCIDT PGVIERVSTL FKGHTELIYG FNMFLPPGYK IEIHSDALGC SVPVVSMPSP 360
361 PGAPTSTGTV HMLTGNSSMS GAGHIAIKTT NAATLTPAAG AGAAAAAAAV AQIQSAGAVN 420
421 LMTHGGASLT QTTIHALQQA TPPQSQSPGG GHVHVSVTNS TAANAVVPGQ PGISVSAHNV 480
481 PQNYSRDRER ATITPTGQVA GAAANVNASA SIVVGGPPTP NSLSELSPHG GAGGGPGAGA 540
541 AQHNLHHIQQ AHQSILLGET GQQNQPVEFN HAITYVNKIK NRFQNQPAKY KKFLEILHDY 600
601 QREQKVMKEG SLNQGKMLTE QEVYTQVAKL FGQDEDLLRE FGQFLPDATN HQSGQYMSKS 660
661 ASVHNDHGKR PTATLSGGAH ITMSSASPAP SGSPLHLGAT TLPQIDKSAH AAAIGNLSAV 720
721 NTSVSIKTYN NNQQQQNHVI GSGLNATRND ILFEKDYHAG LQQQAHQRGA GVGGHHHLAG 780
781 TAAGANIGRP GVGASVMVSY DKEHRNNHHV QKYVGHAPNQ NLTHGHNAKK SPSYGIPSVI 840
841 GSMPHISDNS LDRSSPGISY ATPPLPSGPH GQHNSGSATR RPGDDSLVGH YASGAPPAKR 900
901 PKPYCRDVSF SEASSKCTIS DAAFFDKVRK ALRSPEVYDN FLRCLTLFNQ EIVSKTELLG 960
961 LVSPFLMKFP DLLRWFTDFL GPPSGQPAGG LIDGMPLAAT QRQGGGSSNS SHDRGTSHQS1020
1021 AAEYVQDVDL SSCKRLGASY CALPQSTVPK KCSGRTALCR EVLNDKWVSF PTWASEDSTF1080
1081 VTSRKTQFEE TIYRTEDERF ELDLVIEVNS ATIRVLENLQ KKMSRMSTEE LSKFHLDDHL1140
1141 GGTSQTIHQR AIHRIYGDKS GEIITGMKKN PFVAVPIVLK RLKVKEEEWR EAQKTFNKQW1200
1201 REQNEKYYLK SLDHQAINFK PNDMKALRSK SLFNEIETLY DERHDQEDDA MEPFGPHLVL1260
1261 PYKDKTILDD AANLLIHHVK RQTGIQKQEK QKIKQIIRQF VPDLFFAPRQ PLSDDERDDA1320
1321 FPFLVDDNTK MDVDSPLGRT ESSTRNAKST PSESASPARS NASTSSVTPA GIKKETDDSK1380
1381 ATTGSFAPAS SATASSATPV DDATPSTSSA AAAASAASSS TVSGTEGKPK DDPLSSHKEE1440
1441 GAGSTSSGVA TSPRQAQDTA GAGVDVEIKL EHPADFSNPK LLPPHAHGQR EDESYTLFFA1500
1501 NNNWYLFLRL HAILCDRLHV MYERARLLAI EEERCRVNRR ESTATALRLK PKPEIQVEDY1560
1561 YPTFLDMLKN VLDGNMDSNT FEDTMREMFG IYAYISFTLD KVVSNAVRQL QYCVTERAAL1620
1621 DCVELFATEQ RRGCTGGFCR DAHKTFDREM SYQRKAESIL NEENCFKVYI YKIDCRVTIE1680
1681 LLDSEPEEVD KPAALKAQKF SKYVERLANP ALGGGGNTGR SDSALGNDSV VDGSDIKTEA1740
1741 DEDTAEVNTK A |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
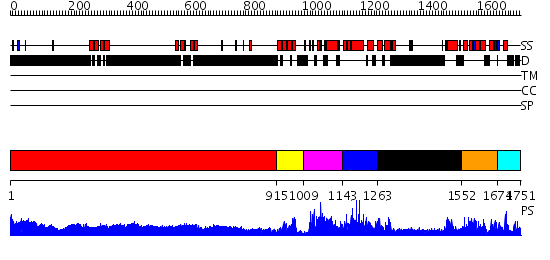
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..914] | 93.522879 | No description for 1y0fB was found. | |
| 2 | View Details | [915..1008] | 1.86 | No description for 2f05A was found. | |
| 3 | View Details | [1009..1142] | 64.376751 | No description for PF08295.3 was found. No confident structure predictions are available. | |
| 4 | View Details | [1143..1262] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1263..1551] | 1.41 | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA | |
| 6 | View Details | [1552..1673] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1674..1751] | 1.019994 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.44 |
Source: Reynolds et al. (2008)