













| General Information: |
|
| Name(s) found: |
CG42347-PB /
FBpp0289206
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 446 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
regulation of cell shape
[IMP]
actin filament organization [IMP] protein amino acid phosphorylation [IGI][NAS] positive regulation of TOR signaling pathway [IGI] |
| Molecular Function: |
ATP binding
[IEA]
protein kinase activity [IGI] protein serine/threonine kinase activity [NAS] myosin light chain kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIYIDDSEPE GVLEPAFPMR DVTINRNVDA HKHYDVLGEV GRGKFGTVYK CRDKANGLQL 60
61 AAKFVPIPKR EDKRNVEREV EIMNSLQHHL IIQLYAAYEY QKMMCVVLEL IEGGELFDRV 120
121 VDDEFVLTER VCRVFIRQVC EAMAFIHGNG IVHLDLKPEN ILVLTQKGNR IKIIDFGLAR 180
181 KFDPDKRLRV LFGTPEFVAP EVVNFDCISY GTDMWSVGVI CYVLISGLSP FMGENDIETM 240
241 SNVTIAKYDF EDECFNGISP ECLDFIAKLL AKDLSTRMTA AECMKHKWLQ QRPATAATAT 300
301 PITKAASAAS KSRLKSVSPV TAPSESSEDS TETIEDEDDE EEVAVQQAKQ KDQQQDEELA 360
361 NLCGDAELEN KELDATKDNL KNFIVRWETH PNSPYVFDVE GNVIAPLSET SYPHPRRTHG 420
421 ADSLSSSRGE YGSQRQRDRH ALIHGK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
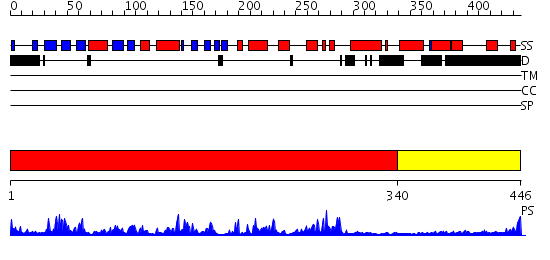
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..339] | 63.30103 | Twitchin, kinase domain | |
| 2 | View Details | [340..446] | 14.045757 | Regulatory apparatus of Calcium Dependent protein kinase from Arabidopsis thaliana |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)