













| General Information: |
|
| Name(s) found: |
Su(var)2-10-PD /
FBpp0087653
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 601 amino acids |
Gene Ontology: |
|
| Cellular Component: |
polytene chromosome, telomeric region
[IDA]
nucleus [IDA][TAS] polytene chromosome [IDA] cytoplasm [IDA][TAS] nuclear lamina [IDA] nucleoplasm [IDA] |
| Biological Process: |
chromosome organization
[IMP][TAS]
hemopoiesis [TAS] chromosome condensation [IMP] regulation of protein catabolic process [TAS] regulation of JAK-STAT cascade [TAS] compound eye development [IMP][TAS] negative regulation of JAK-STAT cascade [NAS] imaginal disc growth [TAS] |
| Molecular Function: |
nucleic acid binding
[IEA]
DNA binding [ISS] zinc ion binding [IEA] DEAD/H-box RNA helicase binding [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRKTRSQTAR TQAENAATSS SPGHQSTSSA PIAVNPFDST KYKECEQMVQ MLRVVELQKI 60
61 LSFLNISFAG RKTDLQSRIL SFLRTNLELL APKVQEVYAQ SVQEQNATLQ YIDPTRMYSH 120
121 IQLPPTVQPN PVGLVGSGQG VQVPGGQMNV VGGAPFLHTH SINSQLPIHP DVRLKKLAFY 180
181 DVLGTLIKPS TLVPRNTQRV QEVPFYFTLT PQQATEIASN RDIRNSSKVE HAIQVQLRFC 240
241 LVETSCDQED CFPPNVNVKV NNKLCQLPNV IPTNRPNVEP KRPPRPVNVT SNVKLSPTVT 300
301 NTITVQWCPD YTRSYCLAVY LVKKLTSTQL LQRMKTKGVK PADYTRGLIK EKLTEDADCE 360
361 IATTMLKVSL NCPLGKMKML LPCRASTCSH LQCFDASLYL QMNERKPTWN CPVCDKPAIY 420
421 DNLVIDGYFQ EVLGSSLLKS DDTEIQLHQD GSWSTPGLRS ETQILDTPSK PAQKVEVISD 480
481 DIELISDDAK PVKRDLSPAQ DEQPTSTSNS ETVDLTLSDS DDDMPLAKRR PPAKQAVASS 540
541 TSNGSGGGQR AYTPAQQPQQ SGGNVFKKPQ KAEELYKRVH LGVWLRPRKP KPESSRRPNV 600
601 G |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
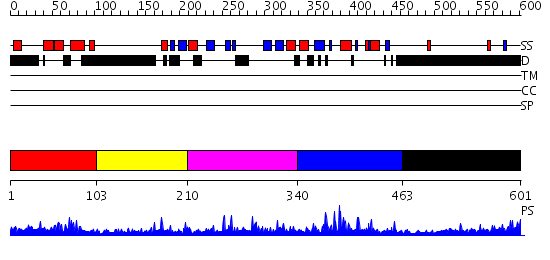
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | 17.0 | Solution structure of human p53 binding domain of PIAS-1 | |
| 2 | View Details | [103..209] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [210..339] | 2.137984 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [340..462] | 3.69897 | brca1 RING domain | |
| 5 | View Details | [463..601] | 8.120996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)