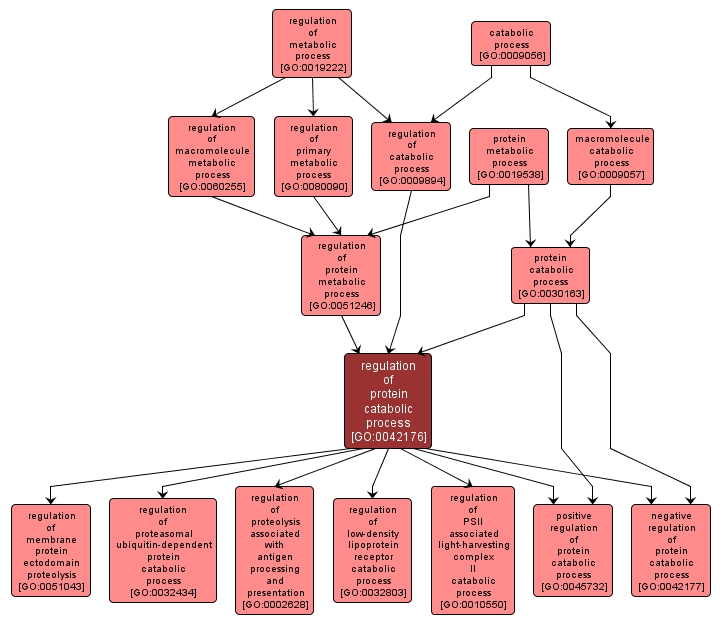
GO TERM SUMMARY
|
| Name: |
regulation of protein catabolic process |
| Acc: |
GO:0042176 |
| Aspect: |
Biological Process |
| Desc: |
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of a protein by the destruction of the native, active configuration, with or without the hydrolysis of peptide bonds. |
Synonyms:
- regulation of protein catabolism
- regulation of protein degradation
- regulation of protein breakdown
|
|

|
INTERACTIVE GO GRAPH
|