













| General Information: |
|
| Name(s) found: |
ACTC_RAT
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Rattus norvegicus |
| Length: | 377 amino acids |
Gene Ontology: |
|
| Cellular Component: |
soluble fraction
[IDA]
cytoskeleton [IEA] cytoplasm [IEA] I band [UNKNOWN][IEA] actin filament [UNKNOWN] cellular_component [ND] actomyosin, actin part [UNKNOWN][IEA] sarcomere [UNKNOWN] |
| Biological Process: |
response to ethanol
[IEP]
heart contraction [UNKNOWN] actomyosin structure organization [UNKNOWN][IEA] skeletal muscle thin filament assembly [UNKNOWN][IEA] response to drug [IEP] apoptosis [UNKNOWN][IEA] cardiac myofibril assembly [UNKNOWN][IEA] actin filament-based movement [UNKNOWN][IEA] cardiac muscle contraction [IDA] cardiac muscle tissue morphogenesis [UNKNOWN][IEA] |
| Molecular Function: |
nucleotide binding
[IEA]
ATPase activity [UNKNOWN][IEA] ATP binding [UNKNOWN][IEA] protein binding [IPI] myosin binding [UNKNOWN][IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCDDEETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA 60
61 QSKRGILTLK YPIEHGIITN WDDMEKIWHH TFYNELRVAP EEHPTLLTEA PLNPKANREK 120
121 MTQIMFETFN VPAMYVAIQA VLSLYASGRT TGIVLDSGDG VTHNVPIYEG YALPHAIMRL 180
181 DLAGRDLTDY LMKILTERGY SFVTTAEREI VRDIKEKLCY VALDFENEMA TAASSSSLEK 240
241 SYELPDGQVI TIGNERFRCP ETLFQPSFIG MESAGIHETT YNSIMKCDID IRKDLYANNV 300
301 LSGGTTMYPG IADRMQKEIT ALAPSTMKIK IIAPPERKYS VWIGGSILAS LSTFQQMWIS 360
361 KQEYDEAGPS IVHRKCF |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
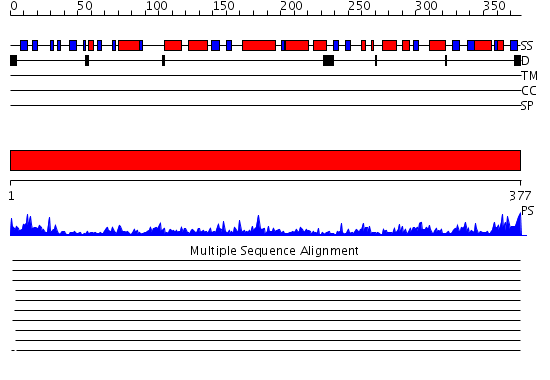
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..377] | 144.0 | Actin |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)