













| General Information: |
|
| Name(s) found: |
SPF30_MOUSE
[Swiss-Prot]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Mus musculus |
| Length: | 238 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
spliceosomal complex [IEA] cellular_component [ND] |
| Biological Process: |
biological_process
[ND]
RNA splicing [IEA] apoptosis [IEA] mRNA processing [IEA] |
| Molecular Function: |
nucleic acid binding
[IEA]
molecular_function [ND] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEDLAKQLA SYKAQLQQVE AALSGNGENE DLLKLKKDLQ EVIELTKDLL STQPSETLAS 60
61 SDSFASTQPT HSWKVGDKCM AVWSEDGQCY EAEIEEIDEE NGTAAITFAV YGNAEVTPLL 120
121 NLKPVEEGRK AKEDSGNKPM SKKEMIAQQR EYKKKKALKK AQRIKELEQE REDQKVKWQQ 180
181 FNNRAYSKNK KGQVKRSIFA SPESVTGKVG VGTCGIADKP MTQYQDTSKY NVRHLMPQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
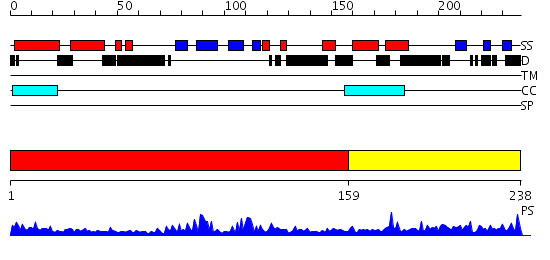
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..158] | 27.69897 | No description for 2hqeA was found. | |
| 2 | View Details | [159..238] | 2.120989 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)