













| General Information: |
|
| Name(s) found: |
TLL1
[HGNC (HUGO)]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 1013 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGLGTLSPRM LVWLVASGIV FYGELWVCAG LDYDYTFDGN EEDKTETIDY KDPCKAAVFW 60
61 GDIALDDEDL NIFQIDRTID LTQNPFGNLG HTTGGLGDHA MSKKRGALYQ LIDRIRRIGF 120
121 GLEQNNTVKG KVPLQFSGQN EKNRVPRAAT SRTERIWPGG VIPYVIGGNF TGSQRAMFKQ 180
181 AMRHWEKHTC VTFIERSDEE SYIVFTYRPC GCCSYVGRRG NGPQAISIGK NCDKFGIVVH 240
241 ELGHVIGFWH EHTRPDRDNH VTIIRENIQP GQEYNFLKME PGEVNSLGER YDFDSIMHYA 300
301 RNTFSRGMFL DTILPSRDDN GIRPAIGQRT RLSKGDIAQA RKLYRCPACG ETLQESNGNL 360
361 SSPGFPNGYP SYTHCIWRVS VTPGEKIVLN FTTMDLYKSS LCWYDYIEVR DGYWRKSPLL 420
421 GRFCGDKLPE VLTSTDSRMW IEFRSSSNWV GKGFAAVYEA ICGGEIRKNE GQIQSPNYPD 480
481 DYRPMKECVW KITVSESYHV GLTFQSFEIE RHDNCAYDYL EVRDGTSENS PLIGRFCGYD 540
541 KPEDIRSTSN TLWMKFVSDG TVNKAGFAAN FFKEEDECAK PDRGGCEQRC LNTLGSYQCA 600
601 CEPGYELGPD RRSCEAACGG LLTKLNGTIT TPGWPKEYPP NKNCVWQVVA PTQYRISVKF 660
661 EFFELEGNEV CKYDYVEIWS GLSSESKLHG KFCGAEVPEV ITSQFNNMRI EFKSDNTVSK 720
721 KGFKAHFFSD KDECSKDNGG CQHECVNTMG SYMCQCRNGF VLHDNKHDCK EAECEQKIHS 780
781 PSGLITSPNW PDKYPSRKEC TWEISATPGH RIKLAFSEFE IEQHQECAYD HLEVFDGETE 840
841 KSPILGRLCG NKIPDPLVAT GNKMFVRFVS DASVQRKGFQ ATHSTECGGR LKAESKPRDL 900
901 YSHAQFGDNN YPGQVDCEWL LVSERGSRLE LSFQTFEVEE EADCGYDYVE LFDGLDSTAV 960
961 GLGRFCGSGP PEEIYSIGDS VLIHFHTDDT INKKGFHIRY KSIRYPDTTH TKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
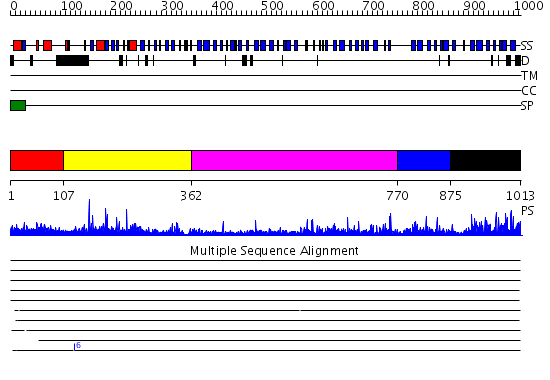
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [107..361] | 40.69897 | STRUCTURE OF ASTACIN AND IMPLICATIONS FOR ACTIVATION OF ASTACINS AND ZINC-LIGATION OF COLLAGENASES | |
| 3 | View Details | [362..769] | 13.69897 | Low density lipoprotein (LDL) receptor YWTD domain; Low density lipoprotein (LDL) receptor, different EGF domains; Ligand-binding domain of low-density lipoprotein receptor | |
| 4 | View Details | [770..874] | 31.0 | Mannose-binding protein associated serine protease 2, MASP2 | |
| 5 | View Details | [875..1013] | 4.39794 | CRYSTAL STRUCTURE OF ACIDIC SEMINAL FLUID PROTEIN (ASFP) AT 1.9 A RESOLUTION: A BOVINE POLYPEPTIDE FROM THE SPERMADHESIN FAMILY |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|