













| General Information: |
|
| Name(s) found: |
SCYL1
[HGNC (HUGO)]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 808 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWFFARDPVR DFPFELIPEP PEGGLPGPWA LHRGRKKATG SPVSIFVYDV KPGAEEQTQV 60
61 AKAAFKRFKT LRHPNILAYI DGLETEKCLH VVTEAVTPLG IYLKARVEAG GLKELEISWG 120
121 LHQIVKALSF LVNDCSLIHN NVCMAAVFVD RAGEWKLGGL DYMYSAQGNG GGPPRKGIPE 180
181 LEQYDPPELA DSSGRVVREK WSADMWRLGC LIWEVFNGPL PRAAALRNPG KIPKTLVPHY 240
241 CELVGANPKV RPNPARFLQN CRAPGGFMSN RFVETNLFLE EIQIKEPAEK QKFFQELSKS 300
301 LDAFPEDFCR HKVLPQLLTA FEFGNAGAVV LTPLFKVGKF LSAEEYQQKI IPVVVKMFSS 360
361 TDRAMRIRLL QQMEQFIQYL DEPTVNTQIF PHVVHGFLDT NPAIREQTVK SMLLLAPKLN 420
421 EANLNVELMK HFARLQAKDE QGPIRCNTTV CLGKIGSYLS ASTRHRVLTS AFSRATRDPF 480
481 APSRVAGVLG FAATHNLYSM NDCAQKILPV LCGLTVDPEK SVRDQAFKAI RSFLSKLESV 540
541 SEDPTQLEEV EKDVHAASSP GMGGAAASWA GWAVTGVSSL TSKLIRSHPT TAPTETNIPQ 600
601 RPTPEGVPAP APTPVPATPT TSGHWETQEE DKDTAEDSST ADRWDDEDWG SLEQEAESVL 660
661 AQQDDWSTGG QVSRASQVSN SDHKSSKSPE SDWSSWEAEG SWEQGWQEPS SQEPPPDGTR 720
721 LASEYNWGGP ESSDKGDPFA TLSARPSTQP RPDSWGEDNW EGLETDSRQV KAELARKKRE 780
781 ERRREMEAKR AERKVAKGPM KLGARKLD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
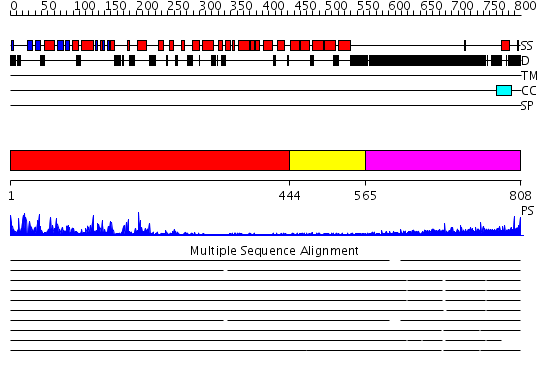
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..443] | 50.69897 | G-protein coupled receptor kinase 2, N-terminal domain; G-protein coupled receptor kinase 2 (beta-adrenergic receptor kinase 1); G-protein coupled receptor kinase 2 | |
| 2 | View Details | [444..564] | 1.44 | No description for 2ie3A was found. | |
| 3 | View Details | [565..808] | 2.331996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)