













| General Information: |
|
| Name(s) found: |
KPNA2
[HGNC (HUGO)]
|
| Description(s) found:
Found 55 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 529 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTNENANTP AARLHRFKNK GKDSTEMRRR RIEVNVELRK AKKDDQMLKR RNVSSFPDDA 60
61 TSPLQENRNN QGTVNWSVDD IVKGINSSNV ENQLQATQAA RKLLSREKQP PIDNIIRAGL 120
121 IPKFVSFLGR TDCSPIQFES AWALTNIASG TSEQTKAVVD GGAIPAFISL LASPHAHISE 180
181 QAVWALGNIA GDGSVFRDLV IKYGAVDPLL ALLAVPDMSS LACGYLRNLT WTLSNLCRNK 240
241 NPAPPIDAVE QILPTLVRLL HHDDPEVLAD TCWAISYLTD GPNERIGMVV KTGVVPQLVK 300
301 LLGASELPIV TPALRAIGNI VTGTDEQTQV VIDAGALAVF PSLLTNPKTN IQKEATWTMS 360
361 NITAGRQDQI QQVVNHGLVP FLVSVLSKAD FKTQKEAVWA VTNYTSGGTV EQIVYLVHCG 420
421 IIEPLMNLLT AKDTKIILVI LDAISNIFQA AEKLGETEKL SIMIEECGGL DKIEALQNHE 480
481 NESVYKASLS LIEKYFSVEE EEDQNVVPET TSEGYTFQVQ DGAPGTFNF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
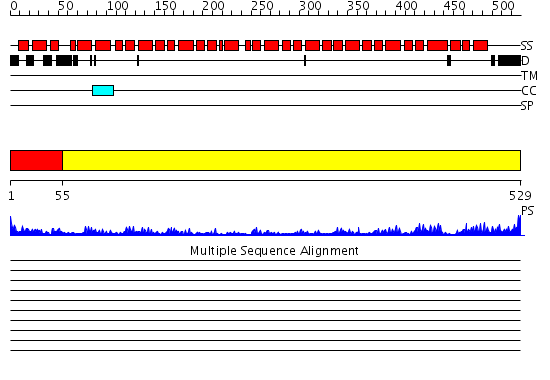
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..54] | 2.49 | Crystal structure of the exportin Cse1p complexed with its cargo ( Kap60p) and RanGTP | |
| 2 | View Details | [55..529] | 103.0 | Mouse Importin alpha- phosphorylated SV40 CN peptide complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)