













| General Information: |
|
| Name(s) found: |
USP11
[HGNC (HUGO)]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 963 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAVAPRLFGG LCFRFRDQNP EVAVEGRLPI SHSCVGCRRE RTAMATVAAN PAAAAAAVAA 60
61 AAAVTEDREP QHEELPGLDS QWRQIENGES GRERPLRAGE SWFLVEKHWY KQWEAYVQGG 120
121 DQDSSTFPGC INNATLFQDE INWRLKEGLV EGEDYVLLPA AAWHYLVSWY GLEHGQPPIE 180
181 RKVIELPNIQ KVEVYPVELL LVRHNDLGKS HTVQFSHTDS IGLVLRTARE RFLVEPQEDT 240
241 RLWAKNSEGS LDRLYDTHIT VLDAALETGQ LIIMETRKKD GTWPSAQLHV MNNNMSEEDE 300
301 DFKGQPGICG LTNLGNTCFM NSALQCLSNV PQLTEYFLNN CYLEELNFRN PLGMKGEIAE 360
361 AYADLVKQAW SGHHRSIVPH VFKNKVGHFA SQFLGYQQHD SQELLSFLLD GLHEDLNRVK 420
421 KKEYVELCDA AGRPDQEVAQ EAWQNHKRRN DSVIVDTFHG LFKSTLVCPD CGNVSVTFDP 480
481 FCYLSVPLPI SHKRVLEVFF IPMDPRRKPE QHRLVVPKKG KISDLCVALS KHTGISPERM 540
541 MVADVFSHRF YKLYQLEEPL SSILDRDDIF VYEVSGRIEA IEGSREDIVV PVYLRERTPA 600
601 RDYNNSYYGL MLFGHPLLVS VPRDRFTWEG LYNVLMYRLS RYVTKPNSDD EDDGDEKEDD 660
661 EEDKDDVPGP STGGSLRDPE PEQAGPSSGV TNRCPFLLDN CLGTSQWPPR RRRKQLFTLQ 720
721 TVNSNGTSDR TTSPEEVHAQ PYIAIDWEPE MKKRYYDEVE AEGYVKHDCV GYVMKKAPVR 780
781 LQECIELFTT VETLEKENPW YCPSCKQHQL ATKKLDLWML PEILIIHLKR FSYTKFSREK 840
841 LDTLVEFPIR DLDFSEFVIQ PQNESNPELY KYDLIAVSNH YGGMRDGHYT TFACNKDSGQ 900
901 WHYFDDNSVS PVNENQIESK AAYVLFYQRQ DVARRLLSPA GSSGAPASPA CSSPPSSEFM 960
961 DVN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample pIC145 from april 2005 | Okada, M., et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
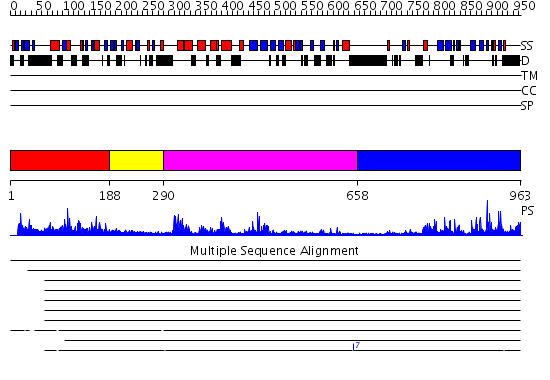
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..187] | 30.39794 | Solution structure of the DUSP domain of hUSP15 | |
| 2 | View Details | [188..289] | 1.51 | Solution Structure of the Ubiquitin-like Domain from Mouse Hypothetical 1700011N24Rik Protein | |
| 3 | View Details | [290..657] | 61.0 | No description for 2gfoA was found. | |
| 4 | View Details | [658..963] | 40.045757 | Ubiquitin carboxyl-terminal hydrolase 7 (USP7, HAUSP) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.56 |
Source: Reynolds et al. (2008)