













| General Information: |
|
| Name(s) found: |
USP16
[HGNC (HUGO)]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 823 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGKKRTKGKT VPIDDSSETL EPVCRHIRKG LEQGNLKKAL VNVEWNICQD CKTDNKVKDK 60
61 AEEETEEKPS VWLCLKCGHQ GCGRNSQEQH ALKHYLTPRS EPHCLVLSLD NWSVWCYVCD 120
121 NEVQYCSSNQ LGQVVDYVRK QASITTPKPA EKDNGNIELE NKKLEKESKN EQEREKKENM 180
181 AKENPPMNSP CQITVKGLSN LGNTCFFNAV MQNLSQTPVL RELLKEVKMS GTIVKIEPPD 240
241 LALTEPLEIN LEPPGPLTLA MSQFLNEMQE TKKGVVTPKE LFSQVCKKAV RFKGYQQQDS 300
301 QELLRYLLDG MRAEEHQRVS KGILKAFGNS TEKLDEELKN KVKDYEKKKS MPSFVDRIFG 360
361 GELTSMIMCD QCRTVSLVHE SFLDLSLPVL DDQSGKKSVN DKNLKKTVED EDQDSEEEKD 420
421 NDSYIKERSD IPSGTSKHLQ KKAKKQAKKQ AKNQRRQQKI QGKVLHLNDI CTIDHPEDSE 480
481 YEAEMSLQGE VNIKSNHISQ EGVMHKEYCV NQKDLNGQAK MIESVTDNQK STEEVDMKNI 540
541 NMDNDLEVLT SSPTRNLNGA YLTEGSNGEV DISNGFKNLN LNAALHPDEI NIEILNDSHT 600
601 PGTKVYEVVN EDPETAFCTL ANREVFNTDE CSIQHCLYQF TRNEKLRDAN KLLCEVCTRR 660
661 QCNGPKANIK GERKHVYTNA KKQMLISLAP PVLTLHLKRF QQAGFNLRKV NKHIKFPEIL 720
721 DLAPFCTLKC KNVAEENTRV LYSLYGVVEH SGTMRSGHYT AYAKARTANS HLSNLVLHGD 780
781 IPQDFEMESK GQWFHISDTH VQAVPTTKVL NSQAYLLFYE RIL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
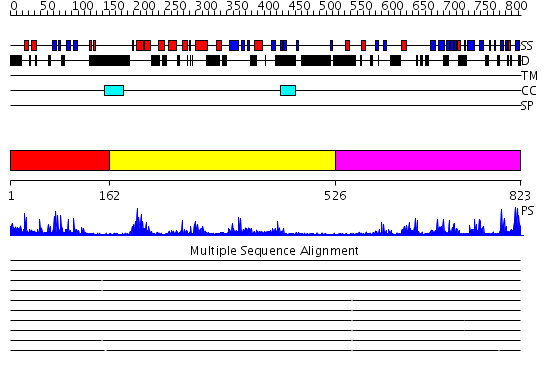
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | 21.09691 | No description for 2i50A was found. | |
| 2 | View Details | [162..525] | 52.69897 | No description for 2gfoA was found. | |
| 3 | View Details | [526..823] | 34.39794 | Ubiquitin carboxyl-terminal hydrolase 7 (USP7, HAUSP) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)