













| General Information: |
|
| Name(s) found: |
YOR338W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 363 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
ascospore formation
[IMP positive regulation of transcription from RNA polymerase II promoter [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLDNMQFHSP APEHPQLNGG INKIPASHKI GYKLNQQVQR LAVVRNNIEE RLNSMESSHG 60
61 QISDSSVVRA IDASIDDFLI PSPPLSPKLR QCPIISQPQL VNVESDHREL IMLTPVWEAG 120
121 LNSQKYNHNT RNFLSQYSFF RDMKTTKRIP NKENRKLKVV KSVVNSEALP KRRRYDRKIK 180
181 RRSRELYEDD GNRSENYDEE SAQEVPVRSV TPIRQVKRSL HTISSPLASQ GVVNNVPKYI 240
241 PSMSWEKLPD YSPPLHTLPN SNNKVLKVEW KGSPMDLNHD PLKQRLHPAE LVLAQILRLP 300
301 CDLYLDSKRR FFLEKVHRFK KGLPFRRTDA QKACRIDVNK ASRLFAAFEK VGWLQDKHFE 360
361 KYL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
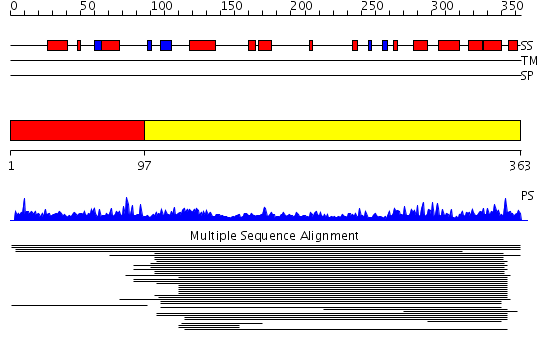
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | 1.007966 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [97..363] | 14.023996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [250..363] | 27.0 | Solution structure of SWIRM domain of mouse transcriptional adaptor 2-like |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)