













| General Information: |
|
| Name(s) found: |
HST3 /
YOR025W
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 447 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
histone deacetylation
[IMP chromatin silencing at telomere [IGI short-chain fatty acid metabolic process [IMP |
| Molecular Function: |
DNA binding
[ISS histone deacetylase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSVSPSPPA SRSGSMCSDL PSSLQTEKLA HIIGLDADDE VLRRVTKQLS RSRRIACLTG 60
61 AGISCNAGIP DFRSSDGLYD LVKKDCSQYW SIKSGREMFD ISLFRDDFKI SIFAKFMERL 120
121 YSNVQLAKPT KTHKFIAHLK DRNKLLRCYT QNIDGLEESI GLTLSNRKLP LTSFSSHWKN 180
181 LDVVQLHGDL KTLSCTKCFQ TFPWSRYWSR CLRRGELPLC PDCEALINKR LNEGKRTLGS 240
241 NVGILRPNIV LYGENHPSCE IITQGLNLDI IKGNPDFLII MGTSLKVDGV KQLVKKLSKK 300
301 IHDRGGLIIL VNKTPIGESS WHGIIDYQIH SDCDNWVTFL ESQIPDFFKT QDQIKKLRQL 360
361 KREASDLRKQ MKAQKDSIGT PPTTPLRTAQ GIDIQGNNEL NTKIKSLNTV KRKILSPENS 420
421 SEEDEEENLD TRKRAKIRPT FGDNQAS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
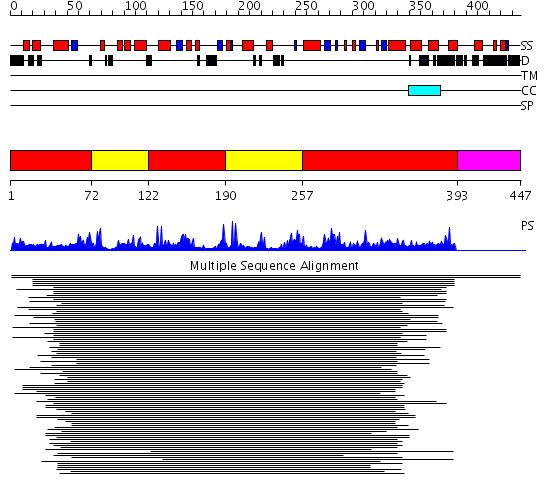
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..71] [122..189] [257..392] |
317.221849 | Sirt2 histone deacetylase | |
| 2 | View Details | [72..121] [190..256] |
317.221849 | Sirt2 histone deacetylase | |
| 3 | View Details | [393..447] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)