













| General Information: |
|
| Name(s) found: |
PFA3 /
YNL326C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 336 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type vacuole membrane
[IDA fungal-type vacuole [IDA |
| Biological Process: |
protein amino acid palmitoylation
[IDA vacuole fusion, non-autophagic [IGI |
| Molecular Function: |
protein-cysteine S-palmitoleyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNDRLSLTSL FPRCLTTCLY IWTAYITLTR IHQIPRWFLA LTIVPTLAVA LYTYYKVIAR 60
61 GPGSPLDFPD LLVHDLKAAE NGLELPPEYM SKRCLTLKHD GRFRVCQVCH VWKPDRCHHC 120
121 SSCDVCILKM DHHCPWFAEC TGFRNQKFFI QFLMYTTLYA FLVLIYTCYE LGTWFNSGSF 180
181 NRELIDFHLL GVALLAVAVF ISVLAFTCFS IYQVCKNQTT IEVHGMRRYR RDLEILNDSY 240
241 GTNEHLENIF DLGSSMANWQ DIMGTSWLEW ILPIETFKYK KSKHTKDEKG LYFNVRPQVQ 300
301 DRLLSSRCLE DQLLRRVTPR PSLEADRASV EIIDAN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
MCD4 PFA3 SHR5 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
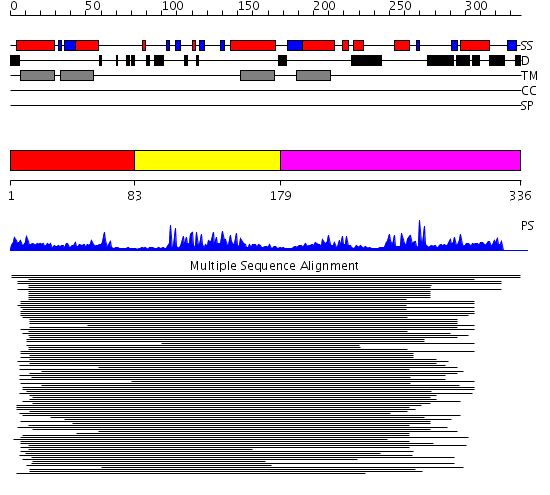
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [83..178] | 37.481486 | DHHC zinc finger domain No confident structure predictions are available. | |
| 3 | View Details | [179..336] | 1.130993 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|