













| General Information: |
|
| Name(s) found: |
NIS1 /
YNL078W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 407 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cellular bud neck [IDA |
| Biological Process: |
regulation of mitosis
[IPI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METYETSIGT QSYPPTLFPP PLGTGGFTTS GYIHALVDST SNSNSNSNTN SNTNSNTNSN 60
61 SDTKIPIVQI SDDSHITHDS FKPYMEYHDA SHLRNRNISK ADQVESTEVM EQFTQWSNYK 120
121 MRSRSPTINA KPIRHTSQRR TDFTSKNELS KFSKNHNFIF HKGFLKRQHS IRREDRQAKV 180
181 RSRFRSKKEL TSVLNYIELE QMDIANVLAS QPVNLHAIRN LTSRDPAVTP IPFLRSQMYA 240
241 TSSRPPYLRN RSISRKLPKS QPGSLPTTMP ATATKTIKQN STTPTTRSVY NKNVGRSNTS 300
301 PSVLYHPKRR GKLNTKSHAR KEQLLLELWR EYLMLVITQR TQLRLTLLCS PGSASNESSV 360
361 CSSNASDLDM SLLSTPSSLF QMAGETKSNP IIIPDSQDDS ILSSDPF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
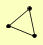
NAP1 NBA1 NIS1 |
| View Details | Krogan NJ, et al. (2006) |

|
NBA1 NIS1 |
| View Details | Gavin AC, et al. (2006) |
|
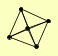
CYS4 NAP1 NBA1 NIS1 SSC1 |
| View Details | Gavin AC, et al. (2002) |
|
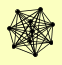
GIN4 HTZ1 KAP114 KCC4 LYS12 NAP1 NBA1 NIS1 SSC1 YRA1 |
| View Details | Ho Y, et al. (2002) |
|
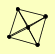
ELP2 ELP3 IKI3 NIS1 RSF2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Hazbun TR, et al. (2003) | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
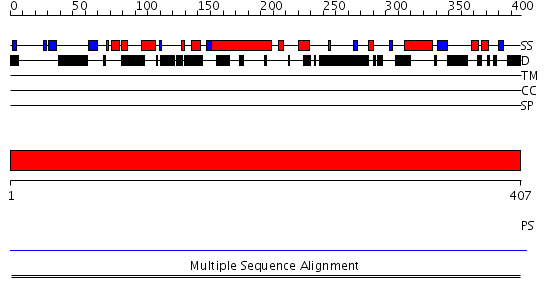
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..407] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)