













| General Information: |
|
| Name(s) found: |
GIS4 /
YML006C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 774 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA |
| Biological Process: |
cellular ion homeostasis
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQKSVRVGDY FDNDDNGLWS WYLTNLRLGD FEELIGNQLK YTLLKRFLNS HFYGDNNISA 60
61 RPNKKILLVS IPENVHEDIS ILEIFLKDYF HLEKLEHIQI SKLTHSHCYN HENHYLLTDN 120
121 LNNFQDPTFL EFASTSWQVQ KNSKALNNNN RNSIPPPTIS SSKASNGKLE SNVSDDQWSN 180
181 INTQTSTATR TNTNTRTLTS PDTVDINVTS VNSQSNNNDT PQDNENEVDE EDATSSIVLN 240
241 FSHSRTVDSK PNRLPKIFPS YTNEDYTPSH SEIMSIDSFA GEDVSSTYPG QDLSLTTARR 300
301 EDESGQDEVE DHYSRVSHDL GDESIDQASY SMESSVSYTS YSSSSNSSSA HYSLSSSSRG 360
361 NPKRENIDHT NATYVSELSS ITSSIDNLTT STTPEEEDNL IHHNYDAQGY GSGEDDGEEV 420
421 YDDEDLSSSD YSVLSILPSI SICDSLGYFR LVLQSILIQD PDTKEIFTAI RQSNNKPTMA 480
481 SVTDDWLLYD SNFSMNNLQI LTLQDLLDIK RSFPKILFYT MVIVTNSGKQ VEEEFKNPNY 540
541 DNREGISKEQ PLDSELSLTN DPQQYFPTAY NNGYNDYIDD EDDEDDGDDA SLSEQSGPQM 600
601 YIPTRMESNV TTAHRSIRTV NSIGEWAFNR HNSVTKIDKS NSNELDNSKT GESTVLSSEP 660
661 HPMTQLSNSN TTSSNFSHSL KTKNSHKPNS KGNNESNSKN ELKKIKSSIN AMSAVERSKS 720
721 LPLPTLLKSL SGIDNPTHAT NKDRKRWKFQ MNRFKNHKNS GSAGTDKSQR CAIM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
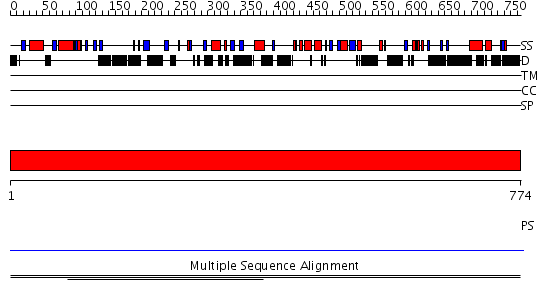
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..774] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [135..389] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [390..536] | 1.00398 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [537..774] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)