













| General Information: |
|
| Name(s) found: |
YLR419W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1435 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKKTKNNSK SSTPVNDVPT TAGKKKAKGK KGQEPEPEDD KRAKQQSNRA KVTSTASWTG 60
61 KLPHTILHET CQKRKWNKVE YDMKKIGDKG FIAIAVLSFT DPKTKETLTA RMNDPTYDKA 120
121 SGKGLVIPQE TPIEARHMAS TIALYRIAYN TNLHMMLPPN HRKTWYALDD FRKDNLKTDE 180
181 KRINKLFDLD PFKTMVEDRK LKAQREKEQV AQNNQAQKEQ VARTILSSHG GISSSGKDRQ 240
241 ERKVASHKNS HNPSLVRFPK KVWENSIFVD LDESSRQLIE TSLKEKIDWQ AKKISHKNET 300
301 IAENREDLKA KLLTLQFRPK HVEEAMLYKD PLSFLLFNLP EDDLPPFFHK KKGDTKNKVE 360
361 ITNLPLSTRM IVERLTEIGV SSDEALLALQ QNDMNENEAA GFLTREILPT LNSNTNEPVS 420
421 ETESIECWNQ ELESLESIYE GCVMDAKEDS HYTLNLIEKL KIKLKVYRTK NYPASLPGIV 480
481 VSTFDKNYKL PDYIKKQILT RLLHYLQEGN LIGDMLVYHI YEWLKENISK IIDNPGPLIP 540
541 DSDSKGAINK RNISNGKRSI NNSSSRKFTK TTISEDTLSV LREEYTKRIK SSEYKSMQLV 600
601 REQLPAWKKQ KVIIDIINKN EVVLITGETG SGKSTQVVQF ILDFLQKEKG DFGKTKIVCT 660
661 QPRRISAIGL AERVSDERCV TCGEEVGYVI RGVNKTKAST RIKFMTTGVL VRLLQNARTM 720
721 LENTIVVIDE VHERSIDTDL IVTLMKNLLH RVRGMKIVLM SATVNVDLFK KFFPGLATCH 780
781 IEGRTFPITD YFLEDILSDL DFKIKREKAL SYDDDSVDER NNDDQYLKPR ADSKFFTSGQ 840
841 INYDLLCQVV EYVHKRLKAA NDNGSIIVFL PGVGEINKCC NLLANKSNEA DFMVLPLHSA 900
901 LTPEDQKRVF KKYHGKRKVV VSTNIAETSI TIDDCVATID TGRAKSMFYN PKDNTTKLIE 960
961 SFISKAEVKQ RRGRAGRVRE GLSYKLFSKN LYENDMISMP IPEIKRIPLE SLYLSVKAMG1020
1021 IKDVKAFLST ALDAPPLPAL QKAERILTTI GLVDESDKSL TQLGQFISLM PVMDSKHGKL1080
1081 LIYGILFGCT DISVLLVSIL GIGVLPFIGG FENREKIKKL LCKYESRGDL FAVLEIVRDY1140
1141 FKIKDSSIKR KYLRDNLLSY NKINEIKSST AQYYSILKDV GFLPMDYKVG SISDLNRNER1200
1201 NFDILRAILT GAFYPHIARV QLPDVKYLST SSGAVEKDPE AKMIKYWIRS EEYQDKLEEY1260
1261 KTKISQETQK VDLEDLPLPA TRAFIHPSSV LFSTNSVNLE DAKLLSEVDG PISRQSKIPT1320
1321 VVKYPFVLFT TSQVTNKLYL RDLTPTTTLS LLLFGGAISY DIGGTIHSPG IVVDNWLPIR1380
1381 TWCKNGVLIK ELRTQLDEAI RKKLESPDYA KKSQIDNSGA DKTLKIVEKI IASEQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
CDC6 HSF1 YLR419W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
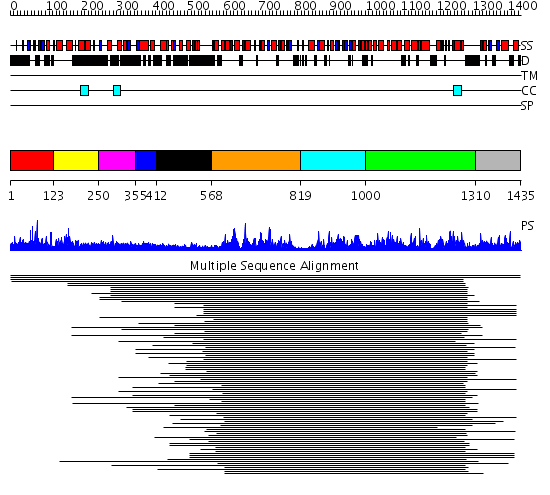
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [123..249] | 1.055994 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [250..354] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [355..411] | 5.207608 | UBA/TS-N domain No confident structure predictions are available. | |
| 5 | View Details | [412..567] | 4.45499 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [568..818] | 46.154902 | Putative DEAD box RNA helicase | |
| 7 | View Details | [819..999] | 46.154902 | Putative DEAD box RNA helicase | |
| 8 | View Details | [1000..1309] | 6.520942 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1310..1435] | 2.05299 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.52 |
Source: Reynolds et al. (2008)