













| General Information: |
|
| Name(s) found: |
YKL091C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 310 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTSILDTYP QICSPNALPG TPGNLTKEQE EALLQFRSIL LEKNYKERLD DSTLLRFLRA 60
61 RKFDINASVE MFVETERWRE EYGANTIIED YENNKEAEDK ERIKLAKMYP QYYHHVDKDG 120
121 RPLYFEELGG INLKKMYKIT TEKQMLRNLV KEYELFATYR VPACSRRAGY LIETSCTVLD 180
181 LKGISLSNAY HVLSYIKDVA DISQNYYPER MGKFYIIHSP FGFSTMFKMV KPFLDPVTVS 240
241 KIFILGSSYK KELLKQIPIE NLPVKYGGTS VLHNPNDKFY YSDIGPWRDP RYIGPEGEIP 300
301 NIFGKFTVTS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
BSP1 PTC7 SEC14 YKL091C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
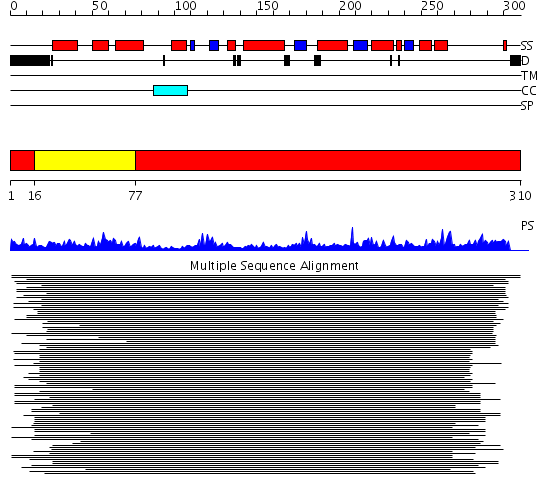
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..15] [77..310] |
1189.522879 | N-terminal domain of phosphatidylinositol transfer protein sec14p; C-terminal domain of phosphatidylinositol transfer protein sec14p | |
| 2 | View Details | [16..76] | 1189.522879 | N-terminal domain of phosphatidylinositol transfer protein sec14p; C-terminal domain of phosphatidylinositol transfer protein sec14p |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)