













| General Information: |
|
| Name(s) found: |
ECM25 /
YJL201W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 599 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
filamentous growth
[IMP response to drug [IMP cellular cell wall organization [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIDINVNNIF FRSYSVDPNS GHAIYVFDST YLPASDEIGD KQVYDLLINA LMDRLVMKLP 60
61 QAPYSLVIFS SGFSQRKISW VYGIKMFAKL PKETKFYLQK IFIVHESFFV RSVYQVISNA 120
121 MNFNFLDSKD SQHDFPSLVH VLDLTSLSEL IDITRLRISL NVYLYDYQIR EHINVPEEYY 180
181 NRLTPLAIRQ YRQLVFDKIF KKLQNDALLC ELIFQKPGNY KKVNIFLDII KRNNYIDLSQ 240
241 WDIYSLASVW LNYFIKNKAK PLIPIELIPL PIVDDLKFTS ETFRKIIKFN QYQDLFMVII 300
301 PFFNRIIAHG ESTKHDSRTL SKALTPALCK EKLSMMTNDR LAIGSRYIKN LLDFFPEIAK 360
361 EISSPPSSVS SSSTIPVLPK PRKSSPTRYS ELGCLTLPRS RSPSPQRSVT SPTYTPVALQ 420
421 NTPVLKPKSS SRNVSSPSFN AKPPLPIKAV TRPQLSLTSN SNTDLALASS STDTLSSPTK 480
481 TPSADSLPLS NSSTDLTISD NIKEMVKDEP AKDKNSVETD IFVQQFESLT LVQNAKIKKF 540
541 DKELQEKKKK NETTSKTADK FSQKGYSDIK ASNKVSRLAA LYEERLQGLQ VMNEMKQRW |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
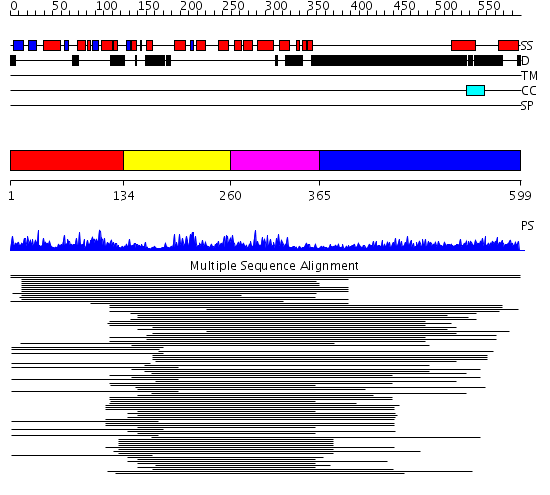
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..133] | 4.028988 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [134..259] | 43.522879 | p50 RhoGAP domain | |
| 3 | View Details | [260..364] | 43.522879 | p50 RhoGAP domain | |
| 4 | View Details | [365..599] | 14.42 | Sec24 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)