













| General Information: |
|
| Name(s) found: |
YJL132W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 750 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane fraction
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSIISSWLLV SIICLTTSIV TKLQAAGVTT HLFYLTRGAP LSLKENYYPW LKAGSFFPDA 60
61 LYSCAPSNKD WSDFAEFTHW PNFLMIAVSY WQQKYGQNDR LRGTHGSLAL KSFLIGVFTH 120
121 QIVDVSWHSL VTDYRMHGLL RVLSETEFDG DIETAHTFLD VMGEFLTLNN VIRDSNNNEN 180
181 WDFLTRSDWK LPREEDLMEI IRNAGLSKEK LSYAELEFCV KRGMAAAISE GYLFRSQRNQ 240
241 LLTNIYSTSP RANDLILNHW LGGQSNLVAM LQRCVPFFET LFHDENTNEA QAEELRLCAN 300
301 LPPVSQKRIN ARPLVSSLKA RKGNSHIVVS PMKSFSDFGT SLTMGKFRED NKDYLAVSAP 360
361 LEDTVGAIYI VPWDILTVAR KEDFSILQPI TAMYGSKVGT YKASDVDYLL VSQPGTCTID 420
421 FYFKGVKILT IKDETTEEAH QLQFAVTGNF YDDKIPDLVV SSPSYGANET GIATFIPGSS 480
481 IISYLTNSDK YQVVDISTFK GVINLDGYPM KIPFQHFGAT IQISDTTDKQ KLIYITCQSL 540
541 GTVFVYSSND LHDLSIPIYY ITKNGVIPAK DSDHVEWHII PSKEHGMFGA AIYSWNFEGM 600
601 SFVAVSQPMF DTVFIYIEKS GQIEFFLKLV LKIKTKSDSI PDEFGSSLLF NDEEKKLYVS 660
661 SPGSFDARGS IWKISMDELL KAGNDPKRKT LLINNLRHLM LINPDKSSKG VSNFGNSMIL 720
721 GPQNHLIVGI PQYGYGNFDH MQLTGRILVL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
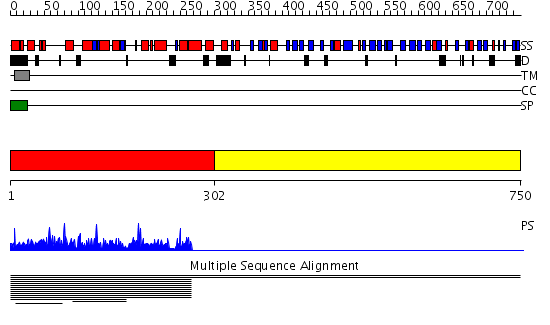
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..301] | 3.006896 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [302..750] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [263..750] | 2.0 | Structural basis for allostery in integrins and binding of ligand-mimetic therapeutics to the platelet receptor for fibrinogen |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|