













| General Information: |
|
| Name(s) found: |
POT1 /
YIL160C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 417 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisomal matrix
[TAS]
|
| Biological Process: |
fatty acid beta-oxidation
[TAS]
|
| Molecular Function: |
acetyl-CoA C-acyltransferase activity
[TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQRLQSIKD HLVESAMGKG ESKRKNSLLE KRPEDVVIVA ANRSAIGKGF KGAFKDVNTD 60
61 YLLYNFLNEF IGRFPEPLRA DLNLIEEVAC GNVLNVGAGA TEHRAACLAS GIPYSTPFVA 120
121 LNRQCSSGLT AVNDIANKIK VGQIDIGLAL GVESMTNNYK NVNPLGMISS EELQKNREAK 180
181 KCLIPMGITN ENVAANFKIS RKDQDEFAAN SYQKAYKAKN EGLFEDEILP IKLPDGSICQ 240
241 SDEGPRPNVT AESLSSIRPA FIKDRGTTTA GNASQVSDGV AGVLLARRSV ANQLNLPVLG 300
301 RYIDFQTVGV PPEIMGVGPA YAIPKVLEAT GLQVQDIDIF EINEAFAAQA LYCIHKLGID 360
361 LNKVNPRGGA IALGHPLGCT GARQVATILR ELKKDQIGVV SMCIGTGMGA AAIFIKE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
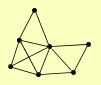
|
CAT2 DCI1 ECI1 FOX2 MLS1 POT1 POX1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
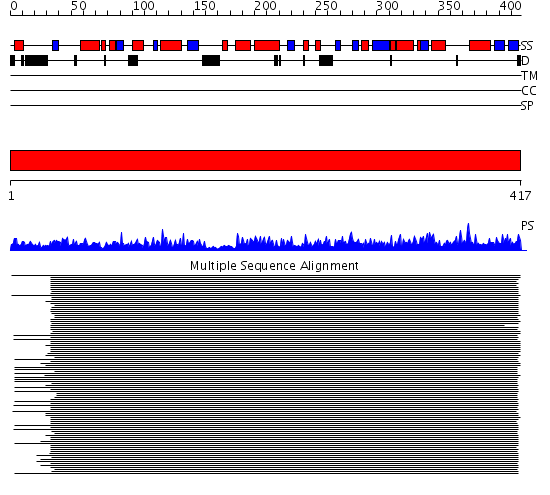
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..417] | 10122.0 | Thiolase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.48 |
Source: Reynolds et al. (2008)