













| General Information: |
|
| Name(s) found: |
DCC1 /
YCL016C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 380 amino acids |
Gene Ontology: |
|
| Cellular Component: |
DNA replication factor C complex
[IPI |
| Biological Process: |
mitotic sister chromatid cohesion
[IMP telomere maintenance [IMP response to DNA damage stimulus [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSINLHSAPE YDPSYKLIQL TPELLDIIQD PVQNHQLRFK SLDKDKSEVV LCSHDKTWVL 60
61 KQRKHSNTVL LMREFVPEQP ITFDETLLFG LSKPYMDVVG FAKTESEFET RETHGELNLN 120
121 SVPIYNGELD FSDKIMKRSS TKVIGTLEEL LENSPCSALE GISKWHKIGG SVKDGVLCIL 180
181 SQDFLFKALH VLLMSAMAES LDLQHLNVED THHAVGKDIE DEFNPYTREI IETVLNKFAV 240
241 QEQEAENNTW RLRIPFIAQW YGIQALRKYV SGISMPIDEF LIKWKSLFPP FFPCDIDIDM 300
301 LRGYHFKPTD KTVQYIAKST LPMDPKERFK VLFRLQSQWD LEDIKPLIEE LNSRGMKIDS 360
361 FIMKYARRKR LGKKTVVTSR |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
CTF8 DCC1 |
| View Details | Krogan NJ, et al. (2006) |
|
AQR1 CTF18 CTF8 DCC1 |
| View Details | Qiu et al. (2008) |
|
CTF18 DCC1 RFC4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
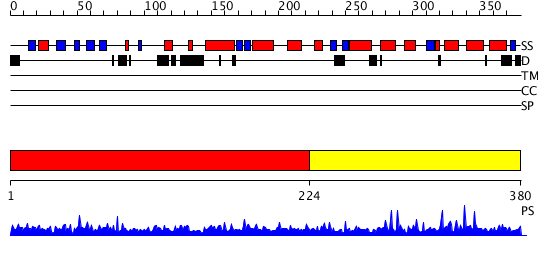
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..223] | 1.007947 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [224..380] | 1.007965 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)