













| General Information: |
|
| Name(s) found: |
PCA1 /
YBR295W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1216 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA |
| Biological Process: |
cellular iron ion homeostasis
[IEP cadmium ion transport [IGI cellular metal ion homeostasis [IMP |
| Molecular Function: |
ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism
[ISS copper ion binding [IMP cadmium-exporting ATPase activity [IMP [NOT]copper-transporting ATPase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKPEKLFSGL GTSDGEYGVV NSENISIDAM QDNRGECHRR SIEMHANDNL GLVSQRDCTN 60
61 RPKITPQECL SETEQICHHG ENRTKAGLDV DDAETGGDHT NESRVDECCA EKVNDTETGL 120
121 DVDSCCGDAQ TGGDHTNESC VDGCCVRDSS VMVEEVTGSC EAVSSKEQLL TSFEVVPSKS 180
181 EGLQSIHDIR ETTRCNTNSN QHTGKGRLCI ESSDSTLKKR SCKVSRQKIE VSSKPECCNI 240
241 SCVERIASRS CEKRTFKGST NVGISGSSST DSLSEKFFSE QYSRMYNRYS SILKNLGCIC 300
301 NYLRTLGKES CCLPKVRFCS GEGASKKTKY SYRNSSGCLT KKKTHGDKER LSNDNGHADF 360
361 VCSKSCCTKM KDCAVTSTIS GTSSSEISRI VSMEPIENHL NLEAGSTGTE HIVLSVSGMS 420
421 CTGCESKLKK SFGALKCVHG LKTSLILSQA EFNLDLAQGS VKDVIKHLSK TTEFKYEQIS 480
481 NHGSTIDVVV PYAAKDFINE EWPQGVTELK IVERNIIRIY FDPKVIGARD LVNEGWSVPV 540
541 SIAPFSCHPT IEVGRKHLVR VGCTTALSII LTIPILVMAW APQLREKIST ISASMVLATI 600
601 IQFVIAGPFY LNALKSLIFS RLIEMDLLIV LSTSAAYIFS IVSFGYFVVG RPLSTEQFFE 660
661 TSSLLVTLIM VGRFVSELAR HRAVKSISVR SLQASSAILV DKTGKETEIN IRLLQYGDIF 720
721 KVLPDSRIPT DGTVISGSSE VDEALITGES MPVPKKCQSI VVAGSVNGTG TLFVKLSKLP 780
781 GNNTISTIAT MVDEAKLTKP KIQNIADKIA SYFVPTIIGI TVVTFCVWIA VGIRVEKQSR 840
841 SDAVIQAIIY AITVLIVSCP CVIGLAVPIV FVIASGVAAK RGVIFKSAES IEVAHNTSHV 900
901 VFDKTGTLTE GKLTVVHETV RGDRHNSQSL LLGLTEGIKH PVSMAIASYL KEKGVSAQNV 960
961 SNTKAVTGKR VEGTSYSGLK LQGGNCRWLG HNNDPDVRKA LEQGYSVFCF SVNGSVTAVY1020
1021 ALEDSLRADA VSTINLLRQR GISLHILSGD DDGAVRSMAA RLGIESSNIR SHATPAEKSE1080
1081 YIKDIVEGRN CDSSSQSKRP VVVFCGDGTN DAIGLTQATI GVHINEGSEV AKLAADVVML1140
1141 KPKLNNILTM ITVSQKAMFR VKLNFLWSFT YNLFAILLAA GAFVDFHIPP EYAGLGELVS1200
1201 ILPVIFVAIL LRYAKI |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
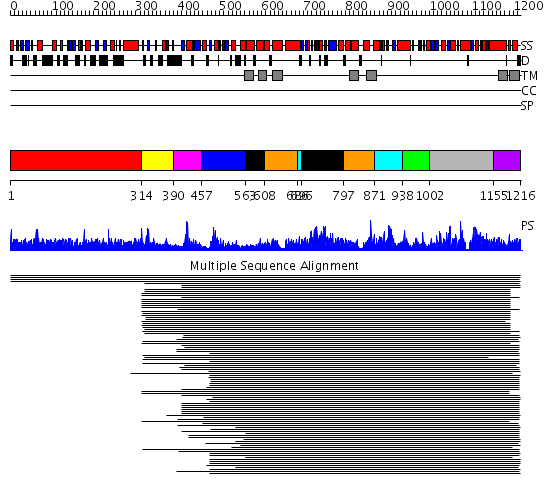
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..313] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [314..389] | 14.522879 | Menkes copper-transporting ATPase | |
| 3 | View Details | [390..456] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [457..562] | 14.39794 | Potential copper-translocating P-type ATPase | |
| 5 | View Details | [563..607] [696..796] |
125.529817 | Calcium ATPase, transduction domain A; Calcium ATPase, catalytic domain P; Calcium ATPase; Calcium ATPase, transmembrane domain M | |
| 6 | View Details | [608..685] [797..870] |
125.529817 | Calcium ATPase, transduction domain A; Calcium ATPase, catalytic domain P; Calcium ATPase; Calcium ATPase, transmembrane domain M | |
| 7 | View Details | [686..695] [871..937] |
125.529817 | Calcium ATPase, transduction domain A; Calcium ATPase, catalytic domain P; Calcium ATPase; Calcium ATPase, transmembrane domain M | |
| 8 | View Details | [938..1001] | 6.39794 | Phosphoserine phosphatase | |
| 9 | View Details | [1002..1154] | 6.39794 | Phosphoserine phosphatase | |
| 10 | View Details | [1155..1216] | 4.447996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|