













| General Information: |
|
| Name(s) found: |
AST1 /
YBL069W
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 429 amino acids |
Gene Ontology: |
|
| Cellular Component: |
extrinsic to membrane
[TAS |
| Biological Process: |
protein targeting to membrane
[IGI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKDILKNQD PKLQAMIVEH SAPAPKEIPM DAPVLKRVAR PLRHVKFIPI KSLIFHTKTG 60
61 PMDFSYEKKI KTPIPKNKIV VRVSNVGLNP VDMKIRNGYT SSIYGEIGLG REYSGVITEV 120
121 GENLNYAWHV GDEVYGIYYH PHLAVGCLQS SILVDPKVDP ILLRPESVSA EEAAGSLFCL 180
181 ATGYNILNKL SKNKYLKQDS NVLINGGTSS VGMFVIQLLK RHYKLQKKLV IVTSANGPQV 240
241 LQEKFPDLAD EMIFIDYLTC RGKSSKPLRK MLEEKKISQY DPVEDKETIL NYNEGKFDVV 300
301 LDFVGGYDIL SHSSSLIHGG GAYVTTVGDY VANYKEDIFD SWDNPSANAR KMFGSIIWSY 360
361 NYTHYYFDPN AKTASANNDW IEQCGDFLKN GTVKCVVDKV YDWKDHKEAF SYMATQRAQG 420
421 KLIMNVEKF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
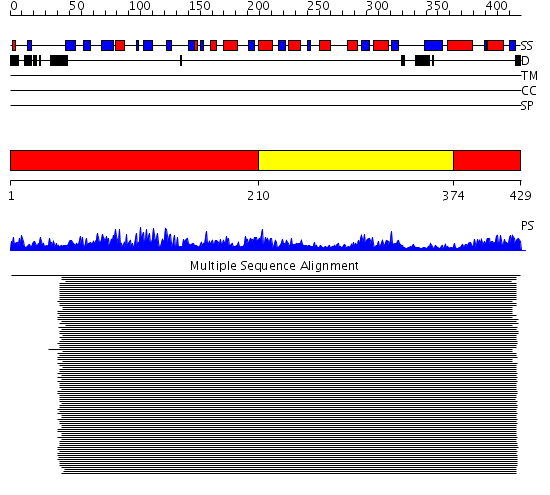
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..209] [374..429] |
121.0 | Alcohol dehydrogenase | |
| 2 | View Details | [210..373] | 121.0 | Alcohol dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)