













| General Information: |
|
| Name(s) found: |
BDH2 /
YAL061W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 417 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRALAYFGKG NIRFTNHLKE PHIVAPDELV IDIEWCGICG TDLHEYTDGP IFFPEDGHTH 60
61 EISHNPLPQA MGHEMAGTVL EVGPGVKNLK VGDKVVVEPT GTCRDRYRWP LSPNVDKEWC 120
121 AACKKGYYNI CSYLGLCGAG VQSGGFAERV VMNESHCYKV PDFVPLDVAA LIQPLAVCWH 180
181 AIRVCEFKAG STALIIGAGP IGLGTILALN AAGCKDIVVS EPAKVRRELA EKMGARVYDP 240
241 TAHAAKESID YLRSIADGGD GFDYTFDCSG LEVTLNAAIQ CLTFRGTAVN LAMWGHHKIQ 300
301 FSPMDITLHE RKYTGSMCYT HHDFEAVIEA LEEGRIDIDR ARHMITGRVN IEDGLDGAIM 360
361 KLINEKESTI KIILTPNNHG ELNREADNEK KEISELSSRK DQERLRESIN EAKLRHT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
BDH1 BDH2 |
| View Details | Krogan NJ, et al. (2006) |

|
BDH1 BDH2 |
| View Details | Gavin AC, et al. (2002) |

|
BDH2 PSE1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
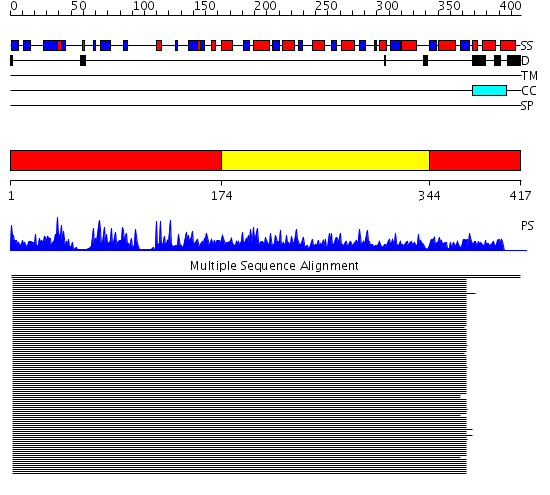
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..173] [344..417] |
463.24799 | Ketose reductase (sorbitol dehydrogenase) | |
| 2 | View Details | [174..343] | 463.24799 | Ketose reductase (sorbitol dehydrogenase) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)