













| General Information: |
|
| Name(s) found: |
BDH1 /
YAL060W
[SGD]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 382 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
glucose catabolic process to butanediol
[IDA |
| Molecular Function: |
(R,R)-butanediol dehydrogenase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRALAYFKKG DIHFTNDIPR PEIQTDDEVI IDVSWCGICG SDLHEYLDGP IFMPKDGECH 60
61 KLSNAALPLA MGHEMSGIVS KVGPKVTKVK VGDHVVVDAA SSCADLHCWP HSKFYNSKPC 120
121 DACQRGSENL CTHAGFVGLG VISGGFAEQV VVSQHHIIPV PKEIPLDVAA LVEPLSVTWH 180
181 AVKISGFKKG SSALVLGAGP IGLCTILVLK GMGASKIVVS EIAERRIEMA KKLGVEVFNP 240
241 SKHGHKSIEI LRGLTKSHDG FDYSYDCSGI QVTFETSLKA LTFKGTATNI AVWGPKPVPF 300
301 QPMDVTLQEK VMTGSIGYVV EAFEEVVRAI HNGDIAMEDC KQLITGKQRI EDGWEKGFQE 360
361 LMDHKESNVK ILLTPNNHGE MK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
BDH1 BDH2 |
| View Details | Krogan NJ, et al. (2006) |

|
BDH1 BDH2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
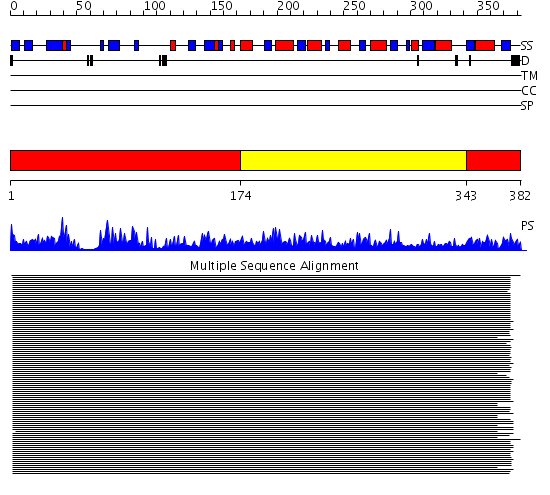
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..173] [343..382] |
388.154902 | Ketose reductase (sorbitol dehydrogenase) | |
| 2 | View Details | [174..342] | 388.154902 | Ketose reductase (sorbitol dehydrogenase) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.56 |
Source: Reynolds et al. (2008)