













| General Information: |
|
| Name(s) found: |
STY8_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 546 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA]
|
| Biological Process: |
metabolic process
[IEA]
protein amino acid phosphorylation [IEA] |
| Molecular Function: |
protein serine/threonine/tyrosine kinase activity
[ISS]
protein kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTIKDESESC GSRAVVASPS QENPRHYRMK LDVYSEVLQR LQESNYEEAT LPDFEDQLWL 60
61 HFNRLPARYA LDVKVERAED VLTHQRLLKL AADPATRPVF EVRSVQVSPR ISADSDPAVE 120
121 EDAQSSHQPS GPGVLAPPTF GSSPNFEAIT QGSKIVEDVD SVVNATLSTR PMHEITFSTI 180
181 DKPKLLSQLT SLLGELGLNI QEAHAFSTVD GFSLDVFVVD GWSQEETDGL RDALSKEILK 240
241 LKDQPGSKQK SISFFEHDKS SNELIPACIE IPTDGTDEWE IDVTQLKIEK KVASGSYGDL 300
301 HRGTYCSQEV AIKFLKPDRV NNEMLREFSQ EVFIMRKVRH KNVVQFLGAC TRSPTLCIVT 360
361 EFMARGSIYD FLHKQKCAFK LQTLLKVALD VAKGMSYLHQ NNIIHRDLKT ANLLMDEHGL 420
421 VKVADFGVAR VQIESGVMTA ETGTYRWMAP EVIEHKPYNH KADVFSYAIV LWELLTGDIP 480
481 YAFLTPLQAA VGVVQKGLRP KIPKKTHPKV KGLLERCWHQ DPEQRPLFEE IIEMLQQIMK 540
541 EVNVVV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
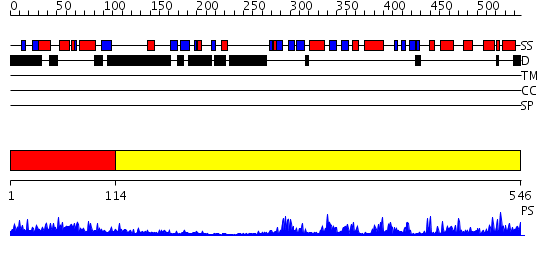
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..113] | 40.69897 | Crystal Structure of the 3 Ig form of FGFR3c in complex with FGF1 | |
| 2 | View Details | [114..546] | 92.154902 | CHICKEN SRC TYROSINE KINASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)