













| General Information: |
|
| Name(s) found: |
gi|24375950
[NCBI NR]
gi|24350938 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 471 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
two-component signal transduction system (phosphorelay)
[ISS regulation of transcription, DNA-dependent [ISS regulation of nitrogen utilization [ISS |
| Molecular Function: |
DNA binding
[ISS two-component response regulator activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRISEQVWIL DDDSSIRWVL EKALQGAKLS TASFAAAESL WQALEISQPR VIVSDIRMPG 60
61 TDGLTLLERL QIHYPHIPVI IMTAHSDLDS AVSAYQAGAF EYLPKPFDID EAISLVERAL 120
121 THATEQSPTP VAQETQVKTP EIIGEAPAMQ EVFRAIGRLS RSSISVLING QSGTGKELVA 180
181 GALHKHSPRK DKPFIALNMA AIPKDLIESE LFGHEKGAFT GAANVRQGRF EQANGGTLFL 240
241 DEIGDMPLDV QTRLLRVLAD GQFYRVGGHS AVQVDVRIIA ATHQDLEQLV LKGGFREDLF 300
301 HRLNVIRIHL PPLSQRREDI PQLASHFLAS AAKEIGVEAK ILTKETAAKL SQLPWPGNVR 360
361 QLENTCRWLT VMASGQEILP QDLPPELLKE PTSINPMAKG SQDWQSALTE WIDQKLSEGN 420
421 SDLLTEVQPA FERILLETAL RHTQGHKQEA AKRLGWGRNT LTRKLKELSM D |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
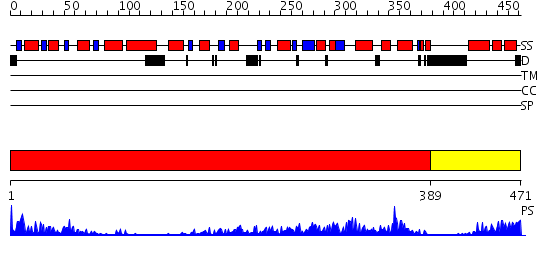
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..388] | 83.69897 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 2 | View Details | [389..471] | 80.522879 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)