













| General Information: |
|
| Name(s) found: |
gi|24374742
[NCBI NR]
gi|24349404 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 446 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
regulation of flagellum assembly
[ISS two-component signal transduction system (phosphorelay) [ISS ciliary or flagellar motility [ISS regulation of transcription, DNA-dependent [ISS |
| Molecular Function: |
DNA binding
[ISS two-component response regulator activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEAKLLLVE DDASLREALL DTLMLAQYDC IDVASGEEAI LALKQHQFDL VISDVQMPGI 60
61 GGLGLLNYLQ QHHPKLPVLL MTAYATIGSA VSAIKLGAVD YLAKPFAPEV LLNQVSRYLP 120
121 LKQNRDQPVV ADEKSLSLLS LAQRVAASDA SVMILGPSGS GKEVLARYIH QHSSRAEEAF 180
181 VAINCAAIPE NMLEATLFGY EKGAFTGAYQ ACPGKFEQAQ GGTLLLDEIS EMDLGLQAKL 240
241 LRVLQEREVE RLGGRKTIKL DVRVLATSNR DLKAVVAAGQ FREDLYYRIN VFPLTWPALN 300
301 QRPADILPLA RHLLTKHAKA LNVVDLPEFD DAACRRLLSH RWPGNVRELD NVVQRALILR 360
361 AGALITANDI IIDAQDVPLT SDDAEYMSEP EGLGEELKAQ EHVIILETLV QCQGSRKLVA 420
421 EKLGISARTL RYKMARMRDM GIQLPS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
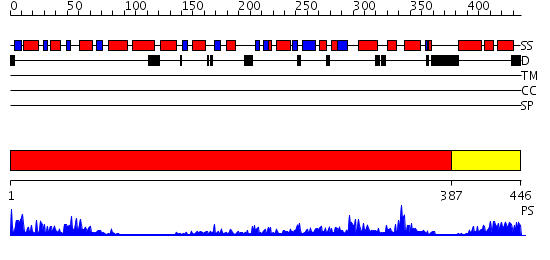
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..386] | 75.0 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 2 | View Details | [387..446] | 70.221849 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)