













| General Information: |
|
| Name(s) found: |
SNF2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1703 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNIPQRQFSN EEVNRCYLRW QHLRNEHGMN APSVPEFIYL TKVLQFAAKQ RQELQMQRQQ 60
61 QGISGSQQNI VPNSSDQAEL PNNASSHISA SASPHLAPNM QLNGNETFST SAHQSPIMQT 120
121 QMPLNSNGGN NMLPQRQSSV GSLNATNFSP TPANNGENAA EKPDNSNHNN LNLNNSELQP 180
181 QNRSLQEHNI QDSNVMPGSQ INSPMPQQAQ MQQAQFQAQQ AQQAQQAQQA QQAQARLQQG 240
241 RRLPMTMFTA EQSELLKAQI TSLKCLVNRK PIPFEFQAVI QKSINHPPDF KRMLLSLSEF 300
301 ARRRQPTDQN NQSNLNGGNN TQQPGTNSHY NNTNTDNVSG LTRNAPLDSK DENFASVSPA 360
361 GPSSVHNAKN GTLDKNSQTV SGTPITQTES KKEENETISN VAKTAPNSNK THTEQNNPPK 420
421 PQKPVPLNVL QDQYKEGIKV VDIDDPDMMV DSFTMPNISH SNIDYQTLLA NSDHAKFTIE 480
481 PGVLPVGIDT HTATDIYQTL IALNLDTTVN DCLDKLLNDE CTESTRENAL YDYYALQLLP 540
541 LQKAVRGHVL QFEWHQNSLL TNTHPNFLSK IRNINVQDAL LTNQLYKNHE LLKLERKKTE 600
601 AVARLKSMNK SAINQYNRRQ DKKNKRLKFG HRLIATHTNL ERDEQKRAEK KAKERLQALK 660
661 ANDEEAYIKL LDQTKDTRIT HLLRQTNAFL DSLTRAVKDQ QKYTKEMIDS HIKEASEEVD 720
721 DLSMVPKMKD EEYDDDDDNS NVDYYNVAHR IKEDIKKQPS ILVGGTLKDY QIKGLQWMVS 780
781 LFNNHLNGIL ADEMGLGKTI QTISLLTYLY EMKNIRGPYL VIVPLSTLSN WSSEFAKWAP 840
841 TLRTISFKGS PNERKAKQAK IRAGEFDVVL TTFEYIIKER ALLSKVKWVH MIIDEGHRMK 900
901 NAQSKLSLTL NTHYHADYRL ILTGTPLQNN LPELWALLNF VLPKIFNSVK SFDEWFNTPF 960
961 ANTGGQDKIE LSEEETLLVI RRLHKVLRPF LLRRLKKDVE KELPDKVEKV VKCKMSALQQ1020
1021 IMYQQMLKYR RLFIGDQNNK KMVGLRGFNN QIMQLKKICN HPFVFEEVED QINPTRETND1080
1081 DIWRVAGKFE LLDRILPKLK ATGHRVLIFF QMTQIMDIME DFLRYINIKY LRLDGHTKSD1140
1141 ERSELLRLFN APDSEYLCFI LSTRAGGLGL NLQTADTVII FDTDWNPHQD LQAQDRAHRI1200
1201 GQKNEVRILR LITTNSVEEV ILERAYKKLD IDGKVIQAGK FDNKSTSEEQ EALLRSLLDA1260
1261 EEERRKKRES GVEEEEELKD SEINEILARN DEEMAVLTRM DEDRSKKEEE LGVKSRLLEK1320
1321 SELPDIYSRD IGAELKREES ESAAVYNGRG ARERKTATYN DNMSEEQWLR QFEVSDDEKN1380
1381 DKQARKQRTK KEDKSEAIDG NGEIKGENID ADNDGPRINN ISAEDRADTD LAMNDDDFLS1440
1441 KKRKAGRPRG RPKKVKLEGS ENSEPPALES SPVTGDNSPS EDFMDIPKPR TAGKTSVKSA1500
1501 RTSTRGRGRG RGRGRGRGRG RGRPPKARNG LDYVRTPAAA TSPIDIREKV AKQALDLYHF1560
1561 ALNYENEAGR KLSDIFLSKP SKALYPDYYM IIKYPVAFDN INTHIETLAY NSLKETLQDF1620
1621 HLIFSNARIY NTEGSVVYED SLELEKVVTK KYCEIMGDNS QLDFTEFDEQ YGTRPLVLPP1680
1681 VVTSSVAESF TDEADSSMTE ASV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
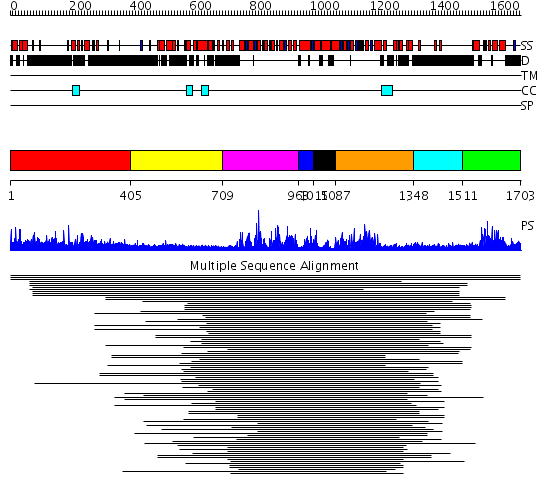
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..404] | 28.176997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [405..708] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [709..962] | 7.221849 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 4 | View Details | [963..1014] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1015..1086] | 7.39794 | Putative DEAD box RNA helicase | |
| 6 | View Details | [1087..1347] | 5.39794 | Nucleotide excision repair enzyme UvrB | |
| 7 | View Details | [1348..1510] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1511..1703] | 87.10721 | GCN5 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)