













| General Information: |
|
| Name(s) found: |
ORC1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 914 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKTLKDLQG WEIITTDEQG NIIDGGQKRL RRRGAKTEHY LKRSSDGIKL GRGDSVVMHN 60
61 EAAGTYSVYM IQELRLNTLN NVVELWALTY LRWFEVNPLA HYRQFNPDAN ILNRPLNYYN 120
121 KLFSETANKN ELYLTAELAE LQLFNFIRVA NVMDGSKWEV LKGNVDPERD FTVRYICEPT 180
181 GEKFVDINIE DVKAYIKKVE PREAQEYLKD LTLPSKKKEI KRGPQKKDKA TQTAQISDAE 240
241 TRATDITDNE DGNEDESSDY ESPSDIDVSE DMDSGEISAD ELEEEEDEEE DEDEEEKEAR 300
301 HTNSPRKRGR KIKLGKDDID ASVQPPPKKR GRKPKDPSKP RQMLLISSCR ANNTPVIRKF 360
361 TKKNVARAKK KYTPFSKRFK SIAAIPDLTS LPEFYGNSSE LMASRFENKL KTTQKHQIVE 420
421 TIFSKVKKQL NSSYVKEEIL KSANFQDYLP ARENEFASIY LSAYSAIESD SATTIYVAGT 480
481 PGVGKTLTVR EVVKELLSSS AQREIPDFLY VEINGLKMVK PTDCYETLWN KVSGERLTWA 540
541 ASMESLEFYF KRVPKNKKKT IVVLLDELDA MVTKSQDIMY NFFNWTTYEN AKLIVIAVAN 600
601 TMDLPERQLG NKITSRIGFT RIMFTGYTHE ELKNIIDLRL KGLNDSFFYV DTKTGNAILI 660
661 DAAGNDTTVK QTLPEDVRKV RLRMSADAIE IASRKVASVS GDARRALKVC KRAAEIAEKH 720
721 YMAKHGYGYD GKTVIEDENE EQIYDDEDKD LIESNKAKDD NDDDDDNDGV QTVHITHVMK 780
781 ALNETLNSHV ITFMTRLSFT AKLFIYALLN LMKKNGSQEQ ELGDIVDEIK LLIEVNGSNK 840
841 FVMEIAKTLF QQGSDNISEQ LRIISWDFVL NQLLDAGILF KQTMKNDRIC CVKLNISVEE 900
901 AKRAMNEDET LRNL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
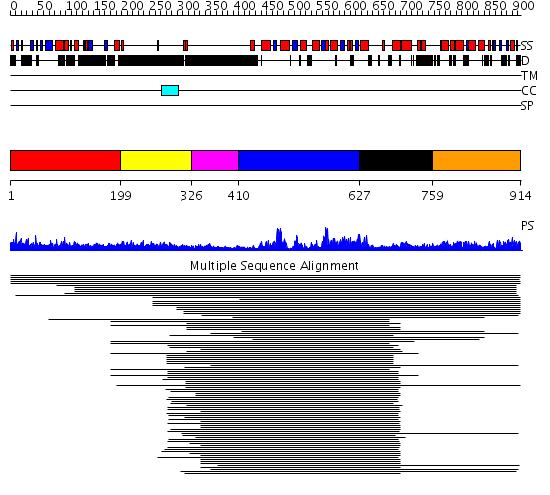
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..198] | 25.045757 | BAH domain No confident structure predictions are available. | |
| 2 | View Details | [199..325] | 2.482996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [326..409] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [410..626] | 51.39794 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 5 | View Details | [627..758] | 51.39794 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 6 | View Details | [759..914] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)